Association of single nucleotide polymorphisms in non‑coding RNAs (miRNA‑100 and MALAT1) with susceptibility to hepatitis B virus infection
- Authors:
- Published online on: July 26, 2023 https://doi.org/10.3892/ije.2023.18
- Article Number: 4
-
Copyright : © Talaat et al. This is an open access article distributed under the terms of Creative Commons Attribution License [CC BY 4.0].
Abstract
Introduction
Hepatitis B infection is a life-threatening liver disease resulting from the hepatitis B virus (HBV) (1). There is high inter-individual variability in the clinical presentation of HBV infection, ranging from self-limited to acute fulminant hepatitis. This can cause chronic liver inflammation leading to cirrhosis and hepatocellular carcinoma (HCC) (2). Despite the presence of effective antiviral therapies and vaccines (3), the mortality rates have increased from 0.8 to 1.4 million from 1990 to 2013(4). Therefore, it is crucial to consider the molecular aspects affecting HBV.
Non-coding RNAs (ncRNAs) are RNA molecules that are not translated into proteins (5). They regulate diverse cellular functions and processes by controlling gene expression (6). High-throughput DNA sequencing and array-based technologies have revolutionized the classification of ncRNAs (7). Among several types of ncRNAs, short ncRNAs, including microRNAs (miRNAs/miRs) and long ncRNAs (lncRNAs), have been considered a standpoint (8).
miRNAs are ~19 to 22 nucleotides (nt) in length (9). To date, >2,000 miRNAs have been registered in the ‘miRbase’ database; however, the functional role of the majority of miRNAs remains unclear; they have emerged as eminent players in human pathophysiological processes (10). miRNAs affect gene expression through various mechanisms, such as de-adenylation, targeting mRNA cleavage and suppression of translation, supporting the evidence that a single miRNA can regulate hundreds of genes; hundreds of miRNAs (11) can also regulate a single gene. An example of miRNAs that have been highly expressed in the liver is miR-100, which is located on chromosome 11 at 11q24.1(12). It promotes HBV protein production, DNA replication and progeny secretion (13).
lncRNAs, the most recent acknowledged class of ncRNAs, are transcripts with lengths >200 nt without protein-coding capacity (14). lncRNAs are messenger RNA (mRNA)-like transcripts, but without stable open reading frames (ORFs). The majority of lncRNAs can regulate gene expression through chromatin modification, transcription and post-transcriptional processing (15). An increasing number of lncRNAs have been characterized in studies, focusing on their roles in regulating gene expression (16,17).
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is one of the most abundant lncRNAs in normal tissues and is highly conserved between humans, also known as nuclear-enriched abundant transcript 2 (NEAT2) (18). It consists of >8,000 nt and is coded by chromosome 11q13. MALAT1 has been reported to regulate gene expression; there is substantial evidence to suggest the vital role of MALAT1 in liver cell proliferation (19,20). Recently, MALAT1 has gained considerable attention due to its association with a number of diseases, also acting as a potential biomarker for the diagnosis, prediction and therapeutic target for numerous types of cancer (21).
Single nucleotide polymorphisms (SNPs) have been suggested to be biological markers for revealing the evolutionary history and common genetic polymorphisms that explain the heritable risk for common diseases (22). SNPs in ncRNAs have been reported to alter their secondary structure or modify expression levels, thereby influencing their regulatory function, contributing to disease development (23). In a previous study, the authors examined the presence of SNP (rs1834306 T/C) in miR-100 in HBV-infected patients and its effect on gene expression (24). In continuation of this, the present study focused on examining SNP rs619586 (A/G) in MALAT1, and its expression level in HBV-infected Egyptian patients compared to other healthy controls. Possible correlations between both miR-100 and MALAT1 in HBV infection were also investigated.
Subjects and methods
Ethics approval
All subjects provided written informed consent for genetic analysis in the present observational prospective case-control study. All methods and analyses were carried out following the guidelines of the Ministry of Health and approved by the Research Ethics Committee for Experimental and Clinical Studies at the Faculty of Pharmacy, University of Cairo, Egypt [BC (1837)].
Patients and study design
A total of 200 subjects; 100 outpatients (70 males and 30 females; under the medical supervision of the National Liver Institute, Menoufia University, Menoufia, Egypt) with an approved diagnosis of HBV infection by enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR) were included in parallel to 100 individuals (64 males and 36 females) with normal liver function test results, no history of hepatic diseases and negative for HBV and hepatitis C virus (HCV) serology, which served as controls. As previously described by Motawi et al (24), viral assessment in all subjects was performed. Patients with HCV infection or other viral or hepatic disorders were excluded from the study. All biochemical investigations included alanine aminotransferase (ALT) and aspartate aminotransferase (AST) activities. According to the manufacturer's instructions, total bilirubin, albumin and creatinine levels were measured in blood for all subjects using a Cobas 6000 analyzer (Roche Diagnostics GmbH).
SNP selection
Based on the data from the HapMap (http://www.hapmap.org); NCBI dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) and miRNAs (http://microrna.sanger.ac.uk) databases, miR100 (rs1834306 T/C) and MALAT1 (rs619586 A/G) SNPs were selected for analyses in the present study.
DNA extraction and SNP genotyping
Genomic DNA was extracted from 5 ml of venous blood samples [collected in ethylene-diamine-tetra-acetic acid (EDTA) sterile vacutainer] from each participant using the GentraPuregene Blood kit (Qiagen GmbH) according to the manufacturer's instructions. A Nanodrop™ 2000/2000c spectrophotometer (Thermo Fisher Scientific, Inc.) was used to assess the purity and the concentration of the extracted DNA. The extracted DNA was applied to 1% agarose gel electrophoresis to confirm its integrity.
miR-100 rs1834306 T/C was analyzed using PCR-sequence-specific primers (PCR-SSP), as previously described by Motawi et al (24). However, MALAT1 polymorphism rs619586 A/G was genotyped by restriction fragment length polymorphism-PCR (RFLP-PCR). The primer sequences of both miR-100 andMALAT1 were designed using Primo SNP 3.4: SNP PCR Primer Design (https://www.changbioscience.com/primo/primosnp.html) and secondly checked using primer blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Table I). A 25 µl PCR reaction mixture contained MyTaq™ Red Mastermix (2X; Meridian Life Science, Inc.), 10 pmoles of each primer and 150 ng DNA. The PCR reaction conditions were as follows: 95˚C for 10 min (one cycle) followed by 35 cycles of 94˚C for the 30 sec, 59˚C for 30 sec, and 72˚C for 1 min, then a final extension step at 72˚C for 7 min. All PCR reactions were performed in a 2720 thermal cycler (Applied Biosystems; Thermo Fisher Scientific, Inc.). A 2% agarose electrophoresis stained with ethidium bromide (10 mg/ml) was used to visualize the PCR product (188 bp) in comparison to the 100 bp DNA ladder (Fermentas; Thermo Fisher Scientific, Inc.) (Fig. 1). The PCR product was digested by the addition of BveI (BspMI) restriction enzymes (Fermentas; Thermo Fisher Scientific, Inc.). The restriction product size was 188bp for the A/A genotype, 121/67 bp for G/G and 188/121/67 bp for A/G. The digestion products were visualized by 3% agarose gel electrophoresis and estimated by comparing with the 50 bp DNA Ladder (Fermentas; Thermo Fisher Scientific, Inc.). In total, 10% of samples were randomly selected to be sequenced to control genotyping quality and validate the results.
Table IPCR primers sequences used for the amplification of miR-100 and MALAT-1 in patients with HBV and the controls. |
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA (from 200 µl plasma samples) was purified using the miRNeasy Mini kit (cat. no. 217004; Qiagen, Inc.) according to the manufacturer's instructions. For the miR-100 expression level, cDNA was prepared using the miScriptII RT kit (cat. no. 218061; Qiagen, Inc.). qPCR was performed using the miScript SYBR-Green PCR kit (cat. no. 218073; Qiagen, Inc.), as previously described in the study by Motawi et al (24).
To determine the transcripts of the gene of interest (MALAT1), RNA was reverse-transcribed using the High-Capacity DNA Reverse Transcription kit (cat no. 4368814; Applied Biosystems; Thermo Fisher Scientific, Inc.). The target cDNA was then amplified using the TaqMan™ Universal Master Mix II (cat no. 4440043; Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control for quantitative normalization. RT-qPCR amplification began with an initial holding period at 50˚C for 2 min, 95˚C for 10 min followed by a PCR program consisting of 40 cycles of 95˚C for 15 sec and 60˚C for 1 min. Differences in the Ct values (Ct) between MALAT1 and GADPH were calculated using the formula ΔΔCt=ΔCt (tested sample)-ΔCt (control sample) to determine the relative expression levels; the fold change in MALAT1 was calculated using the 2-ΔΔCq method (25).
Statistical analysis
All statistical analyses were performed using the clinical Statistical Package for Social Science (SPSS) version 19 (SPSS, Inc.). Data are presented as the mean ± standard deviation/error (SD/SE). An independent paired t-test was applied to compare numerical variables between the patients with HBV and controls for quantitative variables. SNP/STAT was performed using the online tool (http://bioinfo.iconcologia.net/SNP stats). The receiver operating characteristic (ROC) curve analyzed the sensitivity versus specificity of the scoring system. Pearson's correlation analysis was used for correlation analysis. All P-values were two-tailed; A P-value <0.05 was considered to indicate a statistically significant difference.
Results
Demographic and biochemical characteristics of patients with HBV vs. the control subjects
All patients with HBV were found positive for hepatitis B surface antigen (HBsAg) and HBV-DNA. The demographic and biochemical characteristics of the patients with HBV vs. the control subjects have been previously described in the study by Motawi et al (24)
Genetic variation of MALAT1 rs619586 A/G and miR-100 rs1834306 T/C
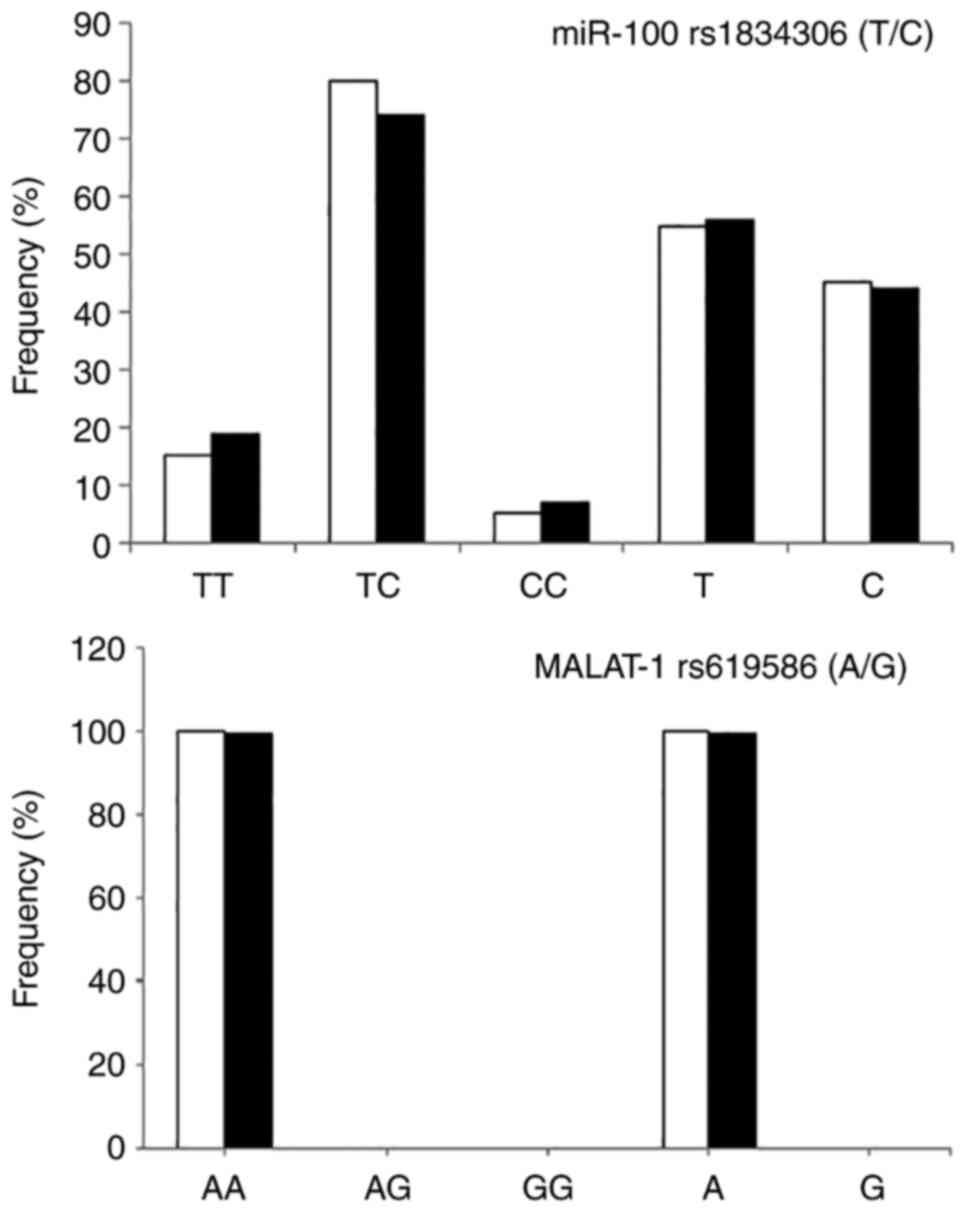
The study of the genotypes of MALAT1 rs619586 A/G revealed the presence of only one genotype: The dominant AA genotype, with a complete disappearance of other genotypes in the two studied groups. On the contrary, all genotypes of miR-100 rs1834306 (T/C) were found, although no statistically significant difference in the genotype distribution between patients with HBV and the normal controls was demonstrated. The genotype and allelic frequency of MALAT1 rs619586 (A/G) and miR-100 rs1834306 (T/C) are presented in Fig. 2.
Expression of miR-100 and MALAT1
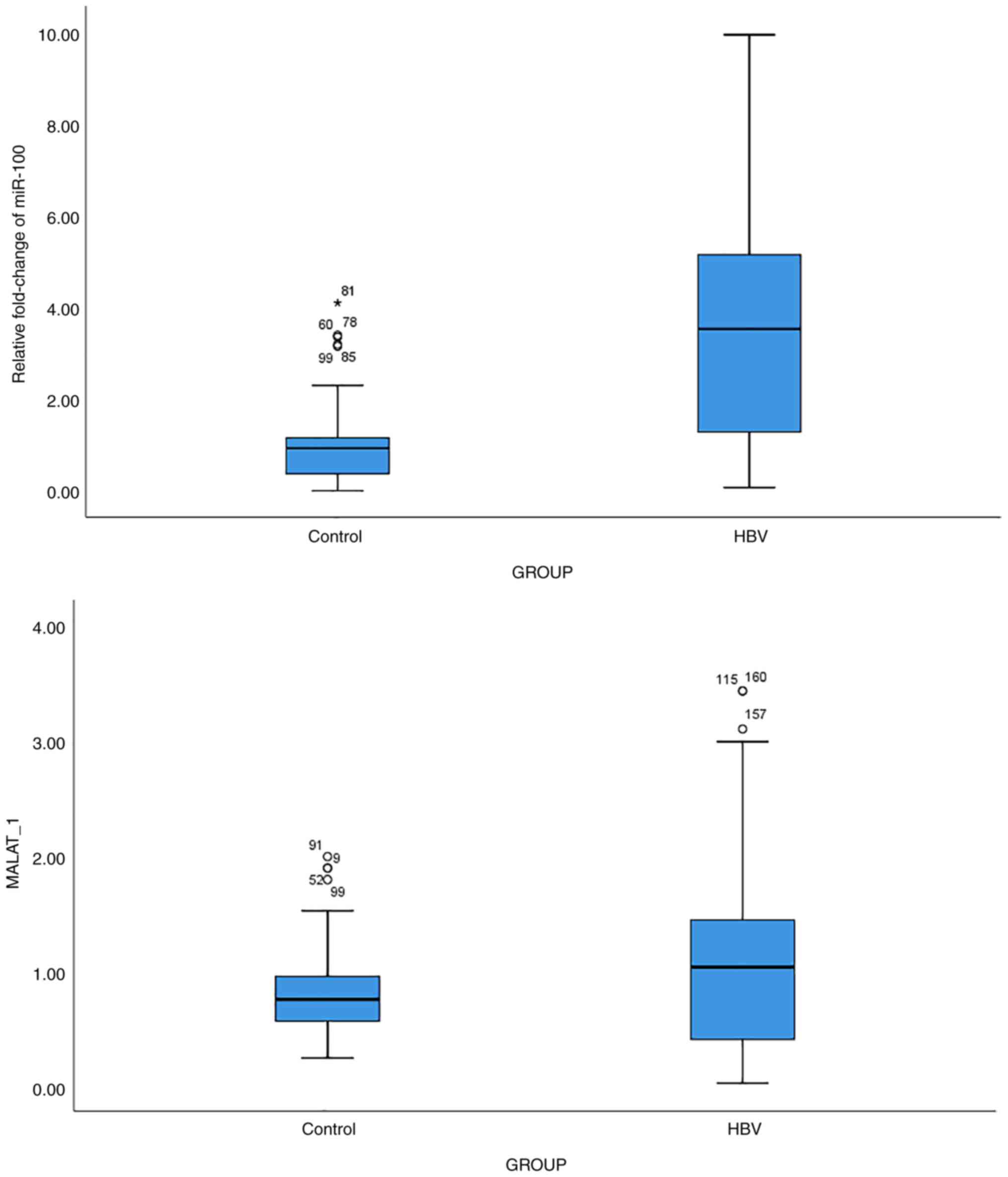
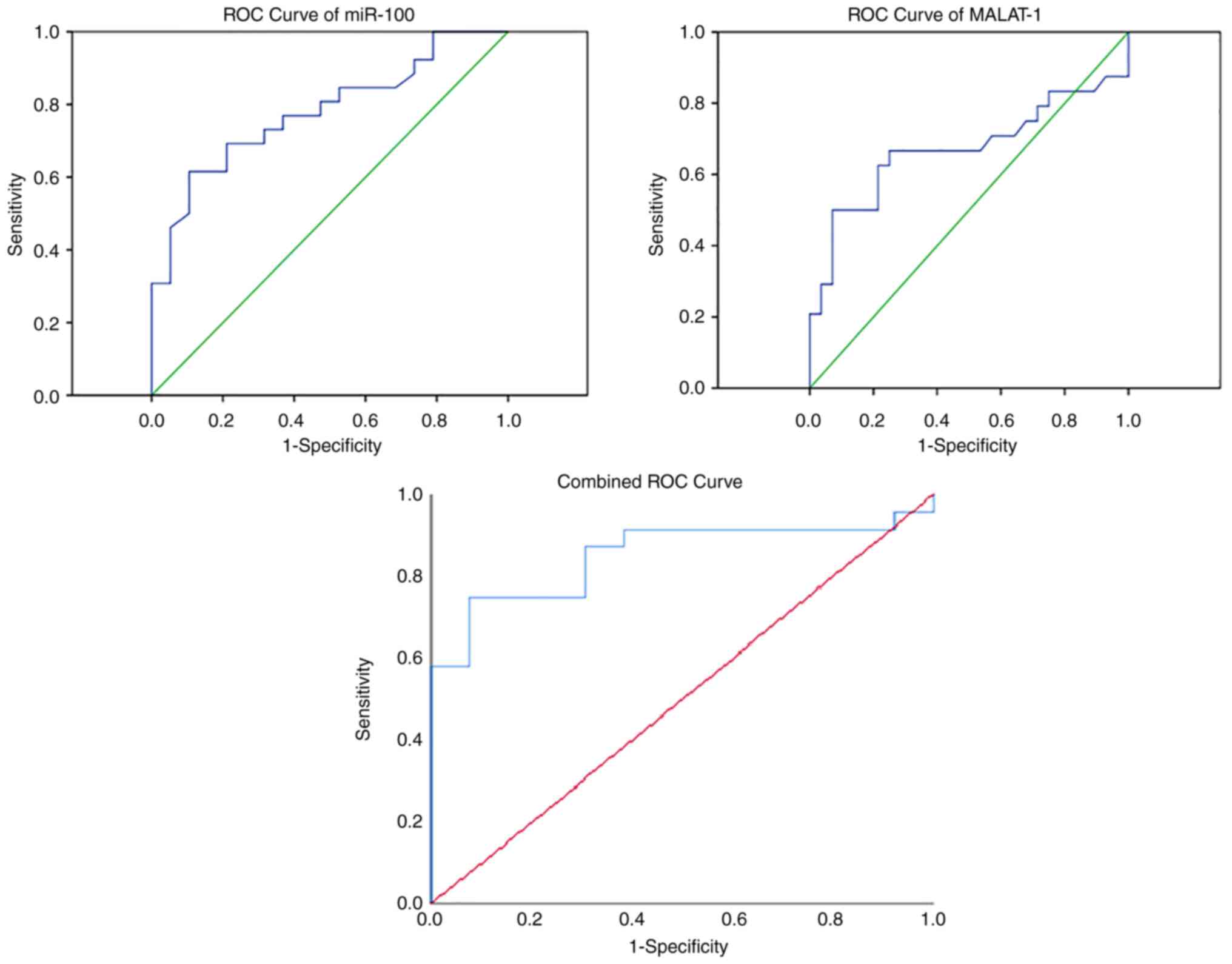
The analysis of the miR-100 and MALAT1 expression levels revealed a significant upregulation ofmiR-100 (P<0.001) and MALAT1 (P<0.001) expression in patients with HBV vs. the controls (Fig. 3). The results of the analysis of the ROC curve for both miR-100 and MALAT1 are summarized in Table II. Combined ROC curve analysis resulted in an improvement in the diagnostic potential of both ncRNAs, leading to 75% sensitivity and 92% specificity (Fig. 4).
Correlations between increased miR-100 and MALAT1 expression levels with some biochemical tests and the viral load in patients HBV vs. the controls
No statistically significant correlation was found between MALAT1 and miR-100 concerning their genotyping or expression level in patients with HBV. A positive correlation between the viral load of HBV, and both MALAT1 expression and (r=0.282 and r2 =0.079; P<0.05) and miR-100 expression (r=0.489 and r2=0.239; P<0.001) was found. In view of the biochemical tests, a positive correlation (r=0.316 and r2=0.099; P<0.05) between MALAT1 expression and the ALT level was detected (Table III).
Table IIICorrelation between biochemical tests and the viral load with miR-100 and MALAT1 expression level. |
Discussion
Viral infections are of global public health concern; HBV is one of the leading causes of mortality (26), and the main obstacle to its treatment is the inability to achieve a full cure for HBV (27). Thus, it is urgent to consider the molecular field affecting HBV. Recent research has devoted ample attention to genetic alterations (28). In this respect, diverse classes of ncRNAs, ranging from miRNAs to lncRNAs, play a crucial role in the epigenetic regulation of gene expression; genome stability also acts as a defense against foreign genetic elements (29).
Over the past decade, an increasing number of scientific studies and research have focused on the pivotal biological functions of one of the key lncRNAs, MALAT1, which was originally discovered as a prognostic marker for lung cancer metastasis, and has been linked to several other human tumor entities (30). Previous studies have revealed that MALAT1 gene polymorphisms are associated with disease susceptibility; for example, the MALAT1 rs619586 has been found to be associated with a decreased risk of developing HCC and colorectal cancer (31-32).
With respect to an SNP of MALAT1 rs619586 A/G in HBV-infected patients vs. the control group, the genotyping results of the present study revealed the dominant appearance of AA genotype in both groups. By contrast, the AG and GG genotypes were lacking. In agreement with these results, Motawi et al (33) demonstrated that the AA genotype was more frequent than AG or GG genotypes in HBV-infected Egyptian patients. An earlier study on the Chinese population reported that the AA genotype was the most frequent with a lack of significance, apart from reporting no significant association between MALAT1 rs619586 SNP and HBV clearance (31). In the Taiwanese population, Yuan et al (34) pointed to an insignificant association between MALAT1 rs619586 and the risk of developing HCC in the HBV-positive subgroup, with higher frequency persistence of the AA genotype over the AG and GG genotypes. However, Wang et al (35) found that tge MALAT1 rs619586 polymorphism decreased the risk of developing HCC under a dominant model, indicating that this SNP has the potential to be a biomarker for HCC risk and prognosis. In general, only a limited number of studies have focused on the genetic variation of MALAT1 rs619586 A/G in HBV infection. In other diseases, the AA genotype is the most dominant genotype, such as in lung cancer (36), congenital heart disease (37), ischemic stroke (38), thyroid carcinoma (39) and recurrent miscarriage (40).
It is not surprising that disease-associated SNPs can alter their gene expression levels (23). Supporting this issue, recent studies have focused (41-46) on the dysregulation of MALAT1 in various diseases. The present study demonstrated a significant upregulation of MALAT1 expression in the plasma of patients with HBV compared to healthy controls. Generally, only a limited number of studies are available to date which quantify MALAT1 expression in patients with HBV. Consistent with the results obtained herein, Konishi et al (41) reported that MALAT1 plasma levels were progressively and significantly elevated in both hepatic disease and patients with HCC. Evidence from other recent studies has reported an increase in the MALAT1 level in HCC tissues (42,43). Multiple lines of evidence have reported the prognostic usefulness of MALAT1 across various types of cancer (44-45), such as being a putative non-invasive biomarker in HCV-induced HCC (46).
In addition to MALAT1, miR-100 is another type of non-protein-coding transcript, miRNA. Previously, it was reported by Motawi et al (25) that miR-100 was significantly upregulated inpatients with HBV, and this elevation is synchronized with the presence of the T allele, suggesting that miR-100 may be considered a potential molecular marker to appraise the prognosis of patients with HBV. As lncRNAs may interact with miRNAs and modulate each other's expression (47), MALAT1 has been described to regulate several miRNAs (48).
The present study hypothesized the presence of correlations between miR-100 and MALAT1. Although no significant correlation was observed, miR-100 and MALAT1 were significantly upregulated in the HBV-infected patients. A positive correlation between the viral load of HBV and both miR-100 and MALAT1 was detected. Both miR-100 and MALAT1 may be regarded as non-invasive molecular markers in HBV infection in the Egyptian population (Fig. 5). Therefore, the present preliminary study focused on two major classes of ncRNAs; miR-100 and MALAT1. Both ncRNAs were upregulated in patients with HBV, and both were found to be positively correlated with the HBV viral load. Accordingly, they may be considered a molecular biomarker in HBV infection. To confirm these findings, further studies with larger sample sizes with other SNPs in both genes are required to clarify the associations between SNPs and their susceptibility to HBV infection in the Egyptian population.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated during and/or analyzed in the current study are available from the corresponding author on reasonable request.
Authors' contributions
The present study was carried out in collaboration between all authors. RMT designed the research and contributed new reagents/analytic tools. SZE provided the patient samples and clinical data. AEM and EAEM performed the experiments. RMT and EAEM analyzed and interpreted the data. SMR and TKM participated in the experimental design and in the writing of the manuscript. RMT, AEM and EAEM confirm the authenticity of the raw data. All authors wrote, and have read and approved the final manuscript.
Ethics approval and consent to participate
All subjects provided written informed consent for genetic analysis in the present observational prospective case-control study. All methods and analyses were carried out followingtheguidelines of the Ministry of Health and approved by the Research Ethics Committee for Experimental and Clinical Studies at the Faculty of Pharmacy, University of Cairo, Egypt [BC (1837)].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Ezbarami ZT, Hassani P, Tafreshi MZ and Majd HA: A qualitative study on individual experiences of chronic hepatitis B patients. Nurs Open. 4:310–318. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Asgari S, Chaturvedi N, Scepanovic P, Hammer C, Semmo N, Giostra E, Müllhaupt B, Angus P, Thompson AJ, Moradpour D and Fellay J: Human genomics of acute liver failure due to hepatitis B virus infection: An exome sequencing study in liver transplant recipients. J Viral Hepat. 26:271–277. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Revill P, Testoni B, Locarnini S and Zoulim F: Global strategies are required to cure and eliminate HBV infection. Nat Rev Gastroenterol Hepatol. 13:239–248. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Alavi M, Grebely J, Hajarizadeh B, Amin J, Larney S, Law MG, George J, Degenhardt L and Dore GJ: Mortality trends among people with hepatitis B and C: A population-based linkage study, 1993-2012. BMC Infect Dis. 18(215)2018.PubMed/NCBI View Article : Google Scholar | |
|
Cao MX, Jiang YP, Tang YL and Liang X: The crosstalk between lncRNA and microRNA in cancer metastasis: Orchestrating the epithelial-mesenchymal plasticity. Oncotarget. 8:12472–12483. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Yamamura S, Imai-Sumida M, Tanaka Y and Dahiya R: Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 75:467–484. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Tripathi R, Chakraborty P and Varadwaj PK: Unraveling long non-coding RNAs through analysis of high-throughput RNA-sequencing data. Noncoding RNA Res. 2:111–118. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Bayoumi AS, Sayed A, Broskova Z, Teoh JP, Wilson J, Su H, Tang YL and Kim IM: Crosstalk between long non-coding RNAs and microRNAs in health and disease. Int J Mol Sci. 17(356)2016.PubMed/NCBI View Article : Google Scholar | |
|
Kohlhapp FJ, Mitra AK, Lengyel E and Peter ME: MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene. 34:5857–5868. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Deng K, Wang H, Guo X and Xia J: The crosstalk between long, non-coding RNAs and microRNAs in gastric cancer. Acta Biochim Biophys Sin (Shanghai). 48:111–116. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Paneru B, Ali A, Al-Tobasei R, Kenney B and Salem M: Crosstalk among lncRNAs, microRNAs and mRNAs in the muscle ‘degradome’ of rainbow trout. Sci Rep. 8(8416)2018.PubMed/NCBI View Article : Google Scholar | |
|
Qin C, Huang RY and Wang ZX: Potential role of miR-100 in cancer diagnosis, prognosis, and therapy. Tumor Biol. 36:1403–1409. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Lin Y, Deng W, Pang J, Kemper T, Hu J, Yin J, Zhang J and Lu M: The microRNA-99 family modulates hepatitis B virus replication by promoting IGF-1R/PI3K/Akt/mTOR/ULK1 signaling-induced autophagy. Cell Microbiol. 19:1–15. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Tan JY and Marques AC: miRNA-mediated crosstalk between transcripts: The missing ‘linc’? Bioessays. 38:295–301. 2016.PubMed/NCBI View Article : Google Scholar | |
|
He RZ, Luo DX and Mo YY: Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 6:6–15. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Bhat SA, Ahmad SM, Mumtaz PT, Malik AA, Dar MA, Urwat U, Shah RA and Ganai NA: Long non-coding RNAs: Mechanism of action and functional utility. Noncoding RNA Res. 1:43–50. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Fernandes JCR, Acuña SM, Aoki JI, Floeter-Winter LM and Muxel SM: Long non-coding RNAs in the regulation of gene expression: Physiology and disease. Noncoding RNA. 5(17)2019.PubMed/NCBI View Article : Google Scholar | |
|
Amodio N, Raimondi L, Juli G, Stamato MA, Caracciolo D, Tagliaferri P and Tassone P: MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol. 11(63)2018.PubMed/NCBI View Article : Google Scholar | |
|
Ren D, Li H, Li R, Sun J, Guo P, Han H, Yang Y and Li J: Novel insight into MALAT-1 in cancer: Therapeutic targets and clinical applications. Oncol Lett. 11:1621–1630. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Sun Y and Ma L: New insights into long non-coding RNA malat1 in cancer and metastasis. Cancers (Basel). 11(216)2019.PubMed/NCBI View Article : Google Scholar | |
|
Zhao M, Wang S, Li Q, Ji Q, Guo P and Liu X: Malat1: A long non-coding RNA highly associated with human cancers. Oncol Lett. 16:19–26. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Fareed M and Afzal M: Single nucleotide polymorphism in genome-wide association of human population: A tool for broad spectrum service. Egypt J Med Hum Genet. 14:123–134. 2013. | |
|
Castellanos-Rubio A and Ghosh S: Disease-associated SNPs in inflammation-related lncRNAs. Front Immunol. 10(420)2019.PubMed/NCBI View Article : Google Scholar | |
|
Motawi TK, Mady AE, Shaheen S, Elshenawy SZ, Talaat RM and Rizk SM: Genetic variation in microRNA-100 (miR-100) rs1834306 T/C associated with Hepatitis B virus (HBV) infection: Correlation with expression level. Infect Genet Evol. 73:444–449. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar | |
|
Fedeli U, Grande E, Grippo F and Frova L: Mortality associated with hepatitis C and hepatitis B virus infection: A nationwide study on multiple causes of death data. World J Gastroenterol. 23:1866–1871. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Lok AS, Zoulim F, Dusheiko G and Ghany MG: Hepatitis B cure: From discovery to regulatory approval. Hepatology. 66:1296–1313. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Ahmadi A, Kaviani S, Yaghmaie M, Pashaiefar H, Ahmadvand M, Jalili M, Alimoghaddam K, Eslamijouybari M and Ghavamzadeh A: Altered expression of MALAT 1 lncRNA in chronic lymphocytic leukemia patients, correlation with cytogenetic findings. Blood Res. 53:320–324. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Frías-Lasserre D and Villagra CA: The importance of ncRNAs as epigenetic mechanisms in phenotypic variation and organic evolution. Front Microbiol. 8(2483)2017.PubMed/NCBI View Article : Google Scholar | |
|
Wu M, Lin Z, Li X, Xin X, An J, Zheng Q, Yang Y and Lu D: HULC cooperates with MALAT1 to aggravate liver cancer stem cells growth through telomere repeat-binding factor 2. Sci Rep. 6(36045)2016.PubMed/NCBI View Article : Google Scholar | |
|
Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J, Zhang Y, Chen J, Shen H and Hu Z: A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. 7(e35145)2012.PubMed/NCBI View Article : Google Scholar | |
|
Zhao K, Jin S, Wei B, Cao S and Xiong Z: Association study of genetic variation of lncRNA MALAT1 with carcinogenesis of colorectal cancer. Cancer Manag Res. 10:6257–6261. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Motawi TMK, El-Maraghy SA, Sabry D and Mehana NA: The expression of long non-coding RNA genes is associated with expression with polymorphisms of HULC rs7763881 and MALAT1 rs619586 in hepatocellular carcinoma and HBV Egyptian patients. J Cell Biochem. 120:14645–14656. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Yuan LT, Chang JH, Lee HL, Yang YC, Su SC, Lin CL, Yang SF and Chien MH: Genetic variants of lncRNA MALAT1 exert diverse impacts on the risk and clinicopathologic characteristics of patients with hepatocellular carcinoma. J Clin Med. 8(1406)2019.PubMed/NCBI View Article : Google Scholar | |
|
Wang BG, Xu Q, Lv Z, Fang XX, Ding HX, Wen J and Yuan Y: Association of twelve polymorphisms in three onco-lncRNa genes with hepatocellular cancer risk and prognosis: A case-control study. World J Gastroenterol. 24:2482–2490. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Gong WJ, Peng JB, Yin JY, Li XP, Zheng W, Xiao L, Tan LM, Xiao D, Chen YX, Li X, et al: Association between well-characterized lung cancer lncRNA polymorphisms and platinum-based chemotherapy toxicity in Chinese patients with lung cancer. Acta Pharmacol Sin. 38:581–590. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Li Q, Zhu W, Zhang B, Wu Y, Yan S, Yuan Y, Zhang H, Li J, Sun K, Wang H and Yu T: The MALAT1 gene polymorphism and its relationship with the onset of congenital heart disease in Chinese. Biosci Rep. 38(BSR20171381)2018.PubMed/NCBI View Article : Google Scholar | |
|
Zhu R, Liu X and He Z: Long non-coding RNA H19 and MALAT1 gene variants in patients with ischemic stroke in a northern Chinese Han population. Mol Brain. 11(58)2018.PubMed/NCBI View Article : Google Scholar | |
|
Wang ML and Liu JX: MALAT1 rs619586 polymorphism functions as a prognostic biomarker in the management of differentiated thyroid carcinoma. J Cell Physiol. 235:1700–1710. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Che D, Yang Y, Xu Y, Fang Z, Pi L, Fu L, Zhou H, Tan Y, Lu Z, Li L, et al: The lncRNA MALAT1 rs619586 G variant confers decreased susceptibility to recurrent miscarriage. Front Physiol. 10(385)2019.PubMed/NCBI View Article : Google Scholar | |
|
Konishi H, Ichikawa D, Yamamoto Y, Arita T, Shoda K, Hiramoto H, Hamada J, Itoh H, Fujita Y, Komatsu S, et al: Plasma level of metastasis-associated lung adenocarcinoma transcript 1 is associated with liver damage and predicts development of hepatocellular carcinoma. Cancer Sci. 107:149–154. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Pan Y, Tong S, Cui R, Fan J, Liu C, Lin Y, Tang J, Xie H, Lin P, Zheng T and Yu X: Long non-coding MALAT1 functions as a competing endogenous RNA to regulate vimentin expression by sponging miR-30a-5p in hepatocellular carcinoma. Cell Physiol Biochem. 50:108–120. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Zhao ZB, Chen F and Bai XF: Long non-coding RNA MALAT1 regulates hepatocellular carcinoma growth under hypoxia via sponging microRNA-200a. Yonsei Med J. 60:727–734. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Wang Z, Katsaros D, Biglia N, Shen Y, Fu Y, Loo LWM, Jia W, Obata Y and Yu H: High expression of long non-coding RNA MALAT1 in breast cancer is associated with poor relapse-free survival. Breast Cancer Res Treat. 171:261–271. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Zhu K, Ren Q and Zhao Y: . LncRNA MALAT1 overexpression promotes proliferation, migration and invasion of gastric cancer by activating the PI3K/AKT pathway. Oncol Lett. 17:5335–5342. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Toraih EA, Ellawindy A, Fala SY, Al Ageeli E, Gouda NS, Fawzy MS and Hosny S: Oncogenic long non-coding RNA MALAT1 and HCV-related hepatocellular carcinoma. Biomed Pharmacother. 102:653–669. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q and Yang M: Silencing of long non-coding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 290:3925–3935. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Liu D, Zhu Y, Pang J, Weng X, Feng X and Guo Y: Knockdown of long non-coding RNA MALAT1 inhibits growth and motility of human hepatoma cells via modulation of miR-195. J Cell Biochem. 119:1368–1380. 2018.PubMed/NCBI View Article : Google Scholar |