Circular RNAs and the regulation of gene expression in diabetic nephropathy (Review)
- Authors:
- Published online on: March 21, 2024 https://doi.org/10.3892/ijmm.2024.5368
- Article Number: 44
-
Copyright: © Benitez et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
1. Introduction
Diabetes mellitus (DM) is a chronic disease characterized by a persistent increase in blood glucose (1), which is caused by dysfunctional insulin secretion, inefficient use of insulin by cells in peripheral tissues due to insulin resistance, or both. The prevalence of DM has increased by >2-fold from 1980 to 2021 (from 4 to 10.5%), and is predicted to rise to 11.3% by 2030 and to 12.2% by 2045 (2). DM is one of the 10 top causes of mortality worldwide, thereby constituting a serious health problem (2). Vascular complications of the disease, such as blindness, kidney failure, heart attack, stroke and lower limb amputation, are the most frequent causes of mortality in patients with diabetes in the short and medium term (3).
DM is recognized as a complex disease caused by a combination of lifestyle and genetic factors (4). Although numerous genetic and non-genetic risk factors interact to trigger DM and its vascular complications, the predictive ability of genetic models remains modest (5). Genetic models are based on combining several gene variants or risk alleles associated with the disease through a genetic risk score (GRS), for instance, variants of genes such as TFCL7, PPARG, KCNJ11, SLC30A8, HHEX, CDKAL1, IGF2BP2 and CDKN2A/B for type 2 diabetes (T2D) (6). Although at least 40 risk alleles have been identified for T2D, the predictive power of the GRS is low due to the small effect size of a number of the genetic loci and thus the small added value of genetic risk compared with clinical risk factors (7,8). These clinical risk factors are phenotype-based and have higher predictive ability, for example, body mass index, dietary habits and glycated hemoglobin level. On the other hand, there is a lack of appropriate models for studies of gene-gene and gene-environment interactions in the risk prediction of DM (5). In addition, the susceptibility to genetic factors related to the onset of macrovascular and microvascular complications in patients with DM does not explain all of the phenotypic variation observed during the disease course (9). Therefore, research on the elements involved in genetic expression control has garnered attention in an to attempt to explain the missing heritability of DM.
The role of non-coding RNA in the regulation of gene expression has been investigated in the last 20 years (10-18). Covalently closed circular RNAs (circRNAs) are an important class of non-coding RNA that have a widespread and specific expression in cells and tissues, which are also stable and highly conserved between species (19-22). These molecules can act as sponges of microRNAs (miRNAs/miRs) or proteins to regulate the transcription of their parental genes or the translation of their targets, and consequently serve an important role in different physiological and pathological processes (23).
Several reports have linked circRNAs to the development and progression of different diseases, including DM and its vascular complications (24-26). CircRNAs can modulate the expression of extracellular matrix (ECM) components, such as fibronectin (FN) and type IV collagen (ColIV), which are implicated in the vascular complications of DM (27).
Although dysregulated expression of circRNAs has been reported in diabetic nephropathy (DN) (28-30), the role of these molecules in the modulation of the advanced glycation end products (AGE)-receptor for AGE (RAGE) pathway by direct or indirect interaction with the proteins that constitute this signaling pathway remains unknown. Identifying these aspects may help to characterize the role of circRNAs in the pathophysiology of DN, and their potential as therapeutic targets or biomarkers of the disease.
The present study aimed to review the current knowledge on circRNAs implicated in DN-related cell processes. Moreover, novel potential interactions that could take place between circRNAs expressed in renal cells under high-glucose concentrations and the transcription factors c-Jun and c-Fos are reported.
2. CircRNA structure, biogenesis and degradation
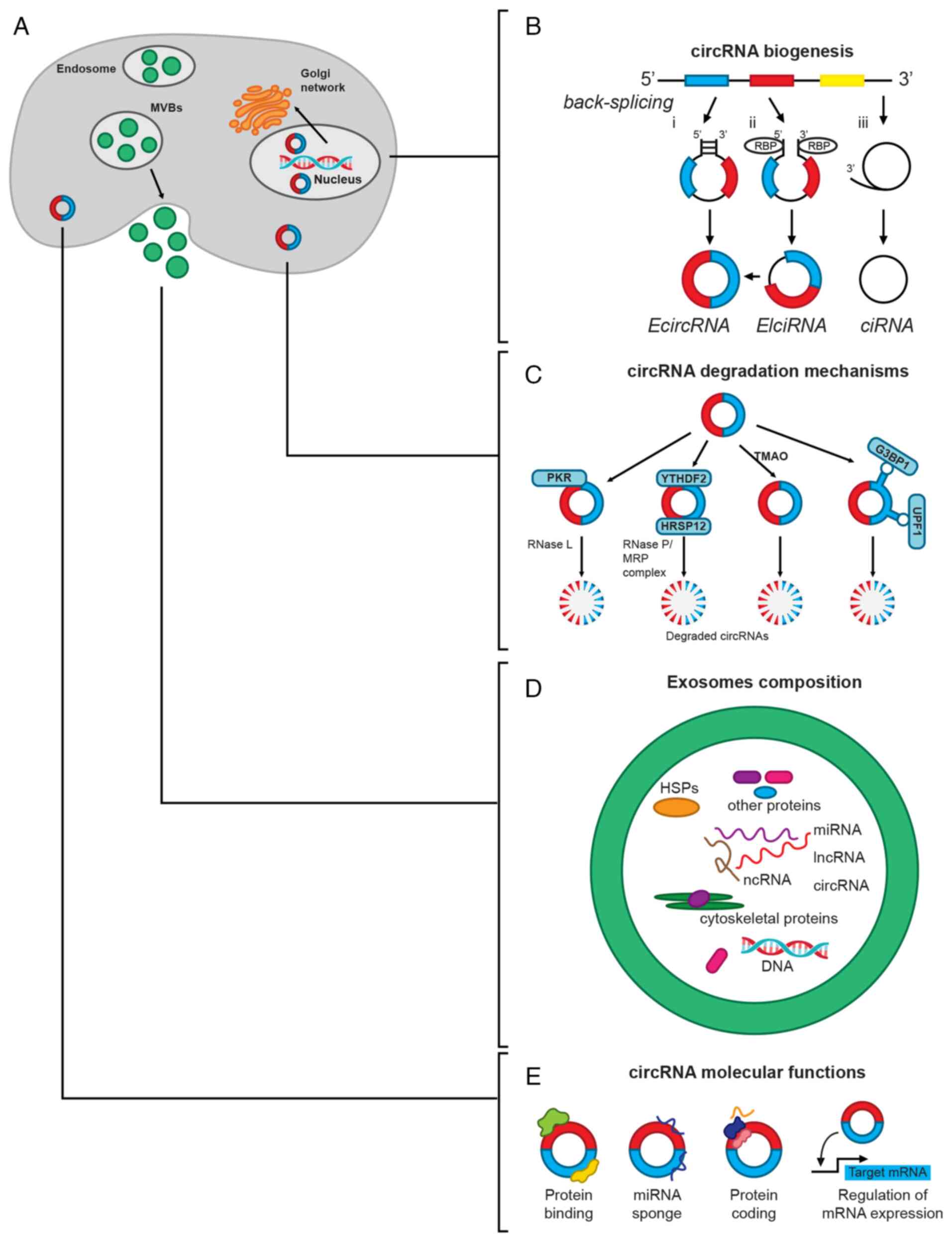
CircRNAs are covalently closed circular single-stranded RNA molecules, which can be in the nucleus, cytoplasm or in exosomes (Fig. 1A). CircRNAs are derived from mRNA backsplicing in which the upstream 5′ splice site of mRNA is linked to a downstream 3′ splice site. The formation of this structure can be driven by direct backsplicing with Arthrobacter luteus elements and inverted repeat complementation, lariat circularization, or it can be mediated by RNA-binding proteins (RBPs) (31,32). Consequently, circRNAs can be generated only from the exon regions of their parental gene [exonic circRNAs (EcircRNAs)], from lariat introns [circular intronic RNAs (ciRNAs)] or from exons with retained introns [exon-intron circRNAs (EIciRNAs)] (Fig. 1B).
According to their origins, different types of circRNAs are located in different cell compartments. EcircRNAs are mainly found in the cytoplasm and are usually the most abundant, constituting ~80% of all the known circRNAs, whereas ciRNAs and EIciRNAs are mainly located in the nuclei. The circRNA parental gene can produce diverse isoforms, but usually only one circRNA isoform is expressed at significant levels (15). CircRNAs are produced in the nucleus and are transported into the cytoplasm via different methods, depending on the length of mature circRNAs and their N6-methyladenosine (m6A) modification status. In human cells, circRNAs are transported into the cytoplasm by URH49 and UAP56, which are two related DEAD box RNA helicases that compose the mRNA processing/nuclear-exporting machinery and regulate gene expression (33). The nuclear export of short circRNAs (<400 nt) is regulated by URH49, whereas the export of long circRNAs (>1,200 nt) is controlled by UAP56. The export mechanism for circRNAs with intermediate lengths has not been completely elucidated (34,35).
Mechanisms for circRNA degradation are mediated by ribonucleases, by the formation of circRNA-protein complexes, by the m6A modification of circRNA or by its packing into exosomes (35) (Fig. 1C and D). Certain circRNAs form DNA:RNA hybrids, displacing a single-stranded DNA (R-loops), which are susceptible to degradation by ribonuclease (RNase) H1. On the other hand, after cell inflammation or viral infection, circRNAs can be degraded by activated RNase L (35) (Fig. 1C).
M6A-modified circRNAs undergo ribonuclease-mediated cleavage via the YTH N6-methyladenosine RBP F2 (YTHDF2)-human heat response protein 12 (HSRP12)-RNase P/MRP axis (36). YTHDF2 is a YTY-domain-containing protein and is classified as a reading protein that recognizes m6A-modified circRNAs, and, using HSRP12 as an adapter, binds to RNase P/MRP to induce degradation of YTHDF2-bound circRNAs (Fig. 1C). Degradation of circRNAs is also mediated by trimethylamine-n-oxide, a byproduct of high sugar and fat diet metabolism of gut microbiota (35) (Fig. 1C).
Argonaute 2 protein, a member of the Argonaute family, also mediates circRNA degradation by recognizing, cleaving and degrading the circRNA-miRNA complex. Other proteins, such as glycine-trytophan protein of 182 kDa, which has an Ago-binding domain and an RNA-recognition motif among other domains, regulate the degradation of certain circRNAs in an Ago-independent manner (35). Another two RBPs, namely up-frameshift protein 1 (UPF1) and Ras-GapSH3 domain-binding protein 1 (G3BP1), which exhibit helicase and GTPase activity, respectively, regulate the degradation of circRNAs depending on the highly folded tridimensional structure present in the majority of these molecules (35) (Fig. 1C).
CircRNAs can also be released from cells packed in exosomes or microvesicles (Fig. 1D), which facilitates their detection and isolation. This is the most important cell mechanism for circRNA removal, and is also a way of regulating intercellular communication through these molecules (35-37). Notably, circRNAs are more abundant than linear mRNAs in exosomes, compared with their abundance in respective parental cells, and they are also more stable, with a half-life of >48 h (38,39). Thus, the presence of large quantities of intact and stable circRNAs in human serum and urine exosomes, alongside their easier detection by liquid biopsies, has suggested the use of circRNAs as a novel diagnostic biomarker and therapeutic target for different diseases, such as cardiovascular diseases, neurological disorders, tumors and renal diseases (40,41).
3. Functions of circRNAs
CircRNAs can regulate gene expression by acting as sponges for miRNAs or proteins (38,39). Other functions include participating as scaffold and cellular translocators (38), as well as regulating the expression of their parental gene (42), the translation of other proteins (43) or their translation to proteins (44) (Fig. 1E). CircRNAs are considered competing/competitive endogenous RNAs because they contain multiple miRNA response elements that competitively bind miRNAs, thus modulating the regulatory function of these molecules (45).
An example of a cytoplasmic circRNA that functions as a miRNA sponge is the cerebellar degeneration related protein 1 antisense transcript/ciRS-7, which has ~70 conserved binding sites for miR-7 and forms a complex with Ago proteins, thus suppressing the degradation of miR-7 target mRNAs (12,39).
CircRNAs also contain binding sites for several RBPs (46-49). The splicing, nuclear export, stability and subcellular localization of mRNAs are all modulated by RBPs (50), such as are Ago proteins, RNA polymerase II and fused in sarcoma (FUS) protein (51). RBPs, besides mediating the backsplicing that drives RNA circularization (Fig. 1B), interact with circRNAs to regulate different processes, such as cell proliferation, apoptosis, cancer cell metastasis, angiogenesis, mRNA translation, energy metabolism and cell differentiation (50).
CircRNAs derived from a specific locus may have binding sites for the protein codified by that locus or another RBP, thus preventing the binding of such proteins to other targets or the mRNA transcribed from the parental gene of the circRNA (52).
CircRNAs may also interact with proteins to modulate their translocation into the nucleus, consequently regulating gene transcription. For example, circRNA_Amotl1 increases the nuclear translocation of STAT3 to regulate the expression of its target genes (53). By contrast, circRNAs can maintain the nuclear retention of the c-Myc protein, increasing its stability and binding affinity to different promoters (52). In addition, circRNAs can act as scaffolds for assembling protein complexes, thus regulating several cellular functions (19). Besides, these RNA molecules can circulate in exosomes in body fluids (40). It has been reported that circRNAs packed in exosomes are regulated by modifying the levels of their target miRNAs in the cells (40).
Previous studies have reported the potential coding properties of circRNAs, albeit at low translational efficiency. This fact is supported by the presence in some circRNAs of an internal ribosomal entry site able to interact with the 40S subunit of the eukaryotic ribosome and an open reading frame ready to be translated into a polypeptide chain. For example, the zinc finger protein 609 circRNA (circRNA_ZNF609) can be translated into a novel ZNF609 protein isoform, which has a potential function during myogenesis (44). CircRNAs may also drive the translation of mRNA by binding to the mature transcript and prevent the start of translation by blocking the interaction between eukaryotic translation initiation factor 4G (EIF4G) and poly-A binding protein (43). It has also been reported that circRNAs may commonly exhibit m6A modifications in response to environmental factors, which promote their protein translation in human cells (27,45).
4. Regulation of the expression of circRNA parental genes
CircRNAs regulate the transcription of their parental genes in several ways (42). One of the methods includes invading the RNA-binding sites in the parental gene, thus blocking the binding of its linear isoform to the corresponding DNA sequence (54). Another mechanism is the conformation of DNA-RNA triple helix (R-loops), which hinders DNA replication (55). The transcription of the parental genes of circRNAs can also be regulated in a cis-acting way (56). For example, circRNAs may interact with U1 small nuclear ribonucleoproteins and RNA polymerase II at the parental gene promoters, thus activating their initiation of transcription. Once transcription is initiated, the expression of circRNAs is increased generating a positive feedback loop. The transcription of the parental gene could be suppressed if the circRNAs interact with transcription factors that promote the expression of such parental genes (52).
CircRNAs may activate intronic enhancers, or induce hypomethylation at the promoter of their parental genes and activate their transcription (57). For example, circRNA_ FECR1 activates the transcription of follicular lymphoma 1 thus regulating the metastatic process of breast cancer (58). Another mechanism of circRNA parental gene regulation is to sequestrate a miRNA that targets a transcription factor, as occurs in the circRNA_STAT3/miR-29a/b/c-3p/glioma-associated oncogene family zinc finger 2 axis, which promotes the progression of hepatoblastoma (59).
The ratio between mRNA-circRNA counterparts may be ≤10:1 due to the competition between backsplicing and linear splicing (60). Consequently, the biogenesis of circRNAs usually provokes a reduction in protein-coding mRNA levels and inhibition of parental gene expression (52).
5. CircRNAs in the pathogenic process of DN
DN is a chronic microvascular complication of DM, which is distinguished by the presence of capillary glomerular circulation damage, which provokes alterations in renal structure and function (61). This disease appears in 30-40% of patients with DM, usually after the first 10 years of disease progression (62). DN is clinically characterized by proteinuria, reduced glomerular filtration rate and high blood pressure, and is the most frequent cause of end-stage renal disease (62). This clinical syndrome is mainly determined by an imbalance between the synthesis and degradation of the ECM components, which provokes their accumulation, as well as the generation of reactive oxygen species (ROS), inflammatory cells recruitment and cytokines release (63). Due to these processes, the glomerular basal membranes and renal tubules thicken, followed by an increase in the volume of the mesangial matrix and glomeruli (64). These events finally lead to inflammation, tubule interstitial renal fibrosis, glomerular sclerosis and tubular atrophy.
Hyperglycemia and oxidative stress are essential mediators in the progression of DN through the formation and intracellular deposition of AGEs (63,65), which are the products of nonenzymatic glycation and oxidation of proteins and lipids (65). AGEs bind to a receptor at the cell surface (i.e. RAGE), which is abundant in kidney podocytes and endothelial cells; thus, the kidneys are considered a major site for AGE clearance (66). The AGE-RAGE interaction triggers multiple signaling cascades that provoke several pathophysiological effects, including cell cycle arrest, apoptosis, increased cell invasion, proliferation and cell migration, and generation of pro-inflammatory cytokines (67,68). For that reason, the increase in AGE formation, the interaction of AGE and RAGE, and the further activation of the associated intracellular signaling pathways have been implicated in the pathogenesis of DN (67,68).
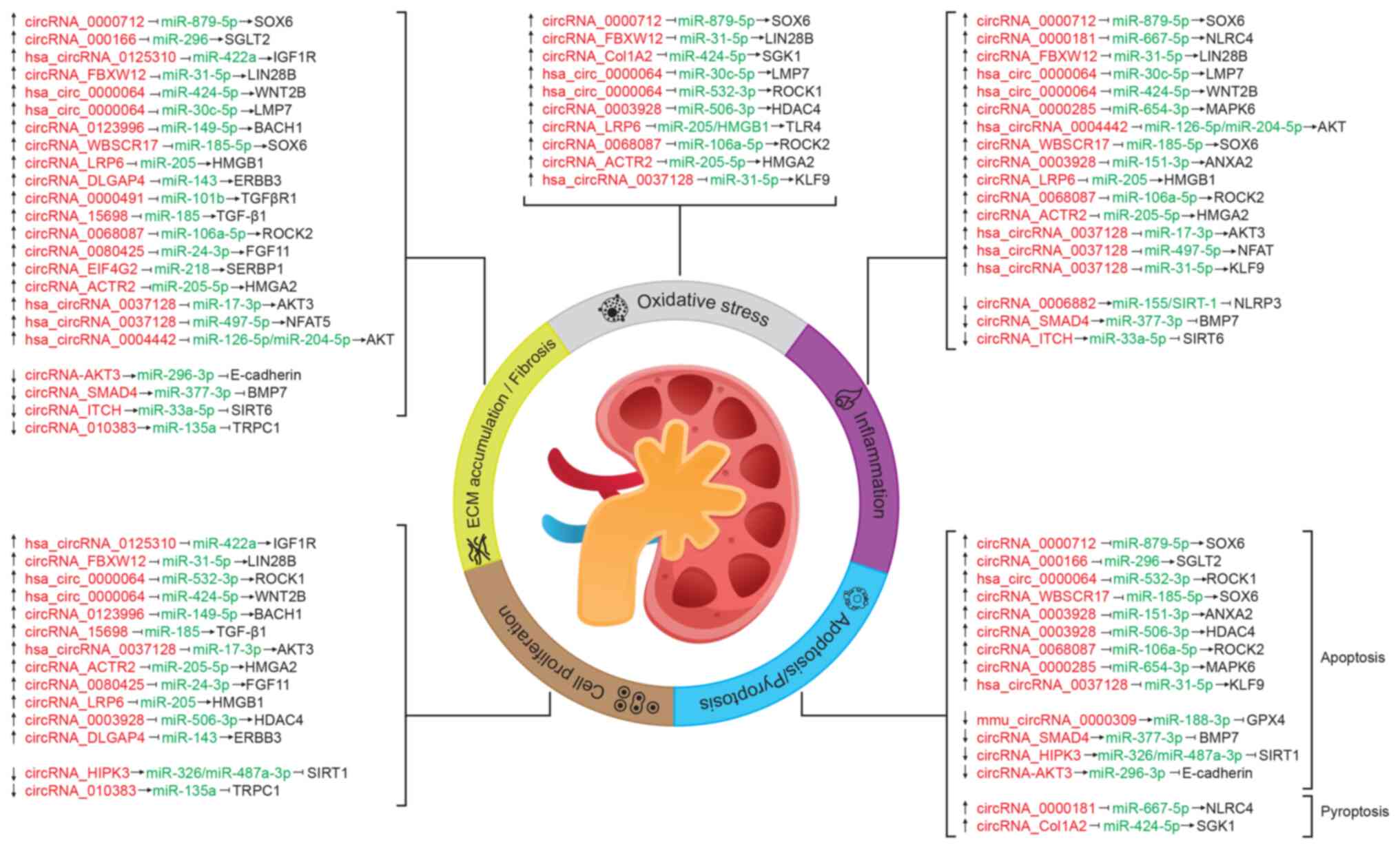
CircRNAs are physiological regulators of multiple intracellular signaling pathways (52); this function is performed indirectly through their interactions with miRNAs or directly with proteins. Several studies have revealed a differential expression profile of circRNAs in kidney cell lines incubated in high glucose, animal models of DN and patients with DN compared with healthy individuals (26-29,69-111). A total of 35 dysregulated circRNAs targeting miRNAs have been identified in these previous studies (Table SI). Of them, ~2/3 are upregulated, one circRNA (circRNA_HIPK3) is upregulated or downregulated depending on the cell type, and the remaining circRNAs are downregulated (Table SI). The target of the identified circRNA-miRNA pairs, as well as the dysregulated axes, have been detected in the majority of cases (28/35; Fig. 2 and Table SI). In most cases, the miRNA target of each circRNA and the mRNA target of the miRNA are suggested through expression studies in the cells or tissues of interest or bioinformatics analysis. Once a negative correlation between the expression of circRNA/miRNA and miRNA/mRNA pairs is verified, the deregulated axes are identified by either silencing or overexpressing the circRNA or miRNA. Then, the expression of the mRNA target, the abundance of its protein, as well as of the effector molecules involved in the molecular processes related to DN are measured (69-104). Fig. 2 shows the best characterized targets and the regulated molecular processes related to DN. The majority of the studied molecules mediate ECM accumulation and fibrosis, whereas circRNAs regulating oxidative stress are less represented (Fig. 2).
Certain circRNAs (namely circRNA_ACTR2, circRNA_LRP6, circRNA_0068087, circRNA_0003928 and circRNA_0000712) simultaneously regulate more than one process, the most common being: Inflammation, oxidative stress and ECM accumulation/fibrosis (Fig. 2 and Table SI). The axes regulated by such circRNAs include as targets transcription factors (circRNA_0000712/miR-879-5p/SOX6), transcriptional regulators [circRNA_ACTR2/miR-205-5p/high mobility group protein HMGI-C (HMGA2), circRNA_LRP6/miR-205/high mobility group protein B1, circRNA_0003928/miR-506-3p/histone deacetylase 4] or protein kinases (circRNA_0068087/miR-106a-5p/Rho-associated protein kinase 2), which modulate different signaling pathways. Notably, hsa_circRNA_0037128 and hsa_circRNA_0000064 mediate oxidative stress, inflammation, apoptosis, cell proliferation and ECM accumulation/fibrosis processes associated with DN, probably because they have more than one target, including transcription factors and protein kinases that are involved in several cell processes (Fig. 2 and Table SI).
Some targets [transcription factor SOX6, RAC-γ serine/threonine-protein kinase (AKT3) and NAD-dependent protein deacetylase sirtuin-1] are regulated by ≥1 circRNA/miRNA pair (Fig. 2 and Table SI), and function in the cellular response to inflammatory, metabolic and oxidative stressors, such as glucose, which suggests that these molecules could be explored as potential therapeutical targets.
Some of the circRNAs that modulate the cell processes associated with DN (Fig. 2 and Table SI) regulate the AGE-RAGE axis and thus may be involved in the pathophysiological events of DN. For example, circRNA_ACTR2 is upregulated in patients with DN, and in both the proximal tubular cell line HK-2 and mesangial cells exposed to high-glucose concentrations (69) (Fig. 2 and Table SI). CircRNA_ACTR2 acts as a sponge for miR-205-5p, which targets HMGA2, a molecule related to the AGE-RAGE pathway (69). HMGA2 is upregulated by AGEs, whereas its knockdown reverses the AGEs-induced epithelial-to-mesenchymal transition of tubular cells associated with DN, and inhibits the high AGEs-induced generation of ROS and the activation of p38 MAPK (112). Consequently, silencing of circRNA_ACTR2 inhibits cell proliferation, inflammatory mediators, ECM deposition and oxidative stress in mesangial cells exposed to a high-glucose concentration (69).
CircRNA_0037128 targets miR-17-3p and is upregulated in kidney tissue of patients with DN, in the mouse mesangial cell line SV40-MES13 when exposed to high glucose levels and in a DN mouse model (70) (Fig. 2 and Table SI). In this previous study, it was demonstrated that the circRNA_0037128/miR17-3p interaction modulated the expression of AKT3, a molecule of the AGE-RAGE axis that promotes cell proliferation and fibrosis (70). The upregulation of circRNA_0037128 can also increase the levels of the pro-inflammatory cytokines TNF-α, IL-1β and IL-6, as well as those of the proteins FN, type I collagen and TGF-β1 in HK-2 tubular cells, while knocking down this circRNA suppresses such effects (71,72). It has been proposed that circRNA_0037128 may regulate these cytokines, and in turn modulate inflammation and fibrosis, through the miR-497-5p/nuclear factor of activated T cells 5 and the miR-31-5p/Krueppel-like factor 9 axes (71,72) (Fig. 2 and Table SI). CircRNA_0037128 is an example of the multiple interactions and regulatory nodes in different cell types that circRNAs can establish. By contrast, circRNA_AKT3 has been shown to be downregulated (Fig. 2 and Table SI), and its overexpression inhibits mouse mesangial cell apoptosis and suppresses ECM accumulation, thus having a protective role in DN via the circRNA_AKT3/miR-296-3p/E-cadherin pathway (73). E-cadherin is a cell adhesion molecule that promotes cell-cell interactions, and allows cohesion between cells and tissue integrity (113). Previous evidence has shown that the AGE-RAGE interaction signal induces tubular epithelial-myofibroblast transdifferentiation, as determined by the loss of the epithelial marker E-cadherin, directly through the dual specificity mitogen-activated protein kinase kinase 1-Ras-extracellular signal-regulated kinase1/2-MAPK pathway (114). Notably, downregulation of circRNA_AKT3 by high-glucose concentrations and AGEs formation may explain the epithelial-to-mesenchymal transition of tubular cells observed in DN.
The actions of circRNA_15698, circRNA_0000491, circRNA_DLGAP4, circRNA_EIF4G2, cirRNA_0000285, hsa_circRNA_0004442 and circRNA_LRP6 are also mediated by molecules of the AGE-RAGE pathway (74-80) (Fig. 2 and Table SI). Most of the remaining circRNAs listed in Table SI regulate molecules involved in inflammation, apoptosis, oxidative stress and fibrosis via other signaling pathway (29,81-104). CircRNA_ANKRD36, hsa_circRNA_0001831 and hsa_ circRNA_0000867 have been shown to be upregulated in blood samples of patients with DN; however, their targets and mechanisms of action remain unknown (105,106) (Table SI). Although miRNAs targeted by hsa_circRNA_0000146 and hsa_circRNA_0000072 have been identified in patients with DN, the evidence only suggests that such circRNAs may be diagnostic markers of the disease (107) (Table SI). Even though several circRNAs have been found to regulate the cellular processes linked to DN, the available data does not support their role in the progression of DN.
It has been recognized that circRNAs may serve a critical role in regulating cellular events by interacting with RBPs (50,115), thus participating in the progression of various diseases (116); however, the role of this type of interaction in DN has not been well explored. A notable example of this type of interaction is the role of circRNA_Amotl1 as an enhancer of cardiomyocyte survival in neonatal human cardiac tissue. Zeng et al (117) reported that circRNA_Amotl1 in primary cardiomyocytes, epithelial and endothelial cells functions as a scaffold of pyruvate dehydrogenase kinase isoform 1 and AKT1, facilitating AKT1 phosphorylation and its nuclear translocation, which reduces apoptosis and enhances cardiac repair. Another example was reported by Stoll et al (118); this previous study demonstrated that intronic circRNA_ci-Ins2/ci-INS binds to TAR DNA-binding protein 43 kDa at the transcriptional level for optimal insulin secretion. Notably, circRNA_ci-Ins2/ci-INS expression is downregulated in pancreatic β-cells of rodent models of diabetes and in patients with T2D (118).
Although it has been demonstrated by global interaction assays that ~8.1% of proteins binding to nucleic acids have a dual function (119) (that is, binding to DNA and RNA), the interaction of circRNA_ci-Ins2/ci-INS with TAR DNA-binding protein 43 kDa in T2D reported by Stoll et al (118) is one of the few reports between circRNAs and proteins with a dual function found in the literature. On the other hand, in type 1 DM, circRNA_PPM1F modulates M1 macrophage activation and inflammation of pancreatic β-cells through the circRNA_ PPM1F/ELAV-like protein 1/protein phosphatase 1F/nuclear factor NF-κB (NF-κB) axis (120).
To the best of our knowledge, only a single report on the interaction of circRNA/protein in DN has been published to date. CircRNA_HIPK3 targets several miRNAs in renal tubular and mesangial cells in rodent models of DN (29,82) (Table SI), although it can also bind proteins. CircRNA_HIPK3 binds FUS and facilitates the enrichment of this protein on the ectodysplasin A2 receptor (EDA2R) promoter; this leads to the upregulation of EDA2R expression and activation of apoptotic signaling in podocytes, which contributes to DN progression (23).
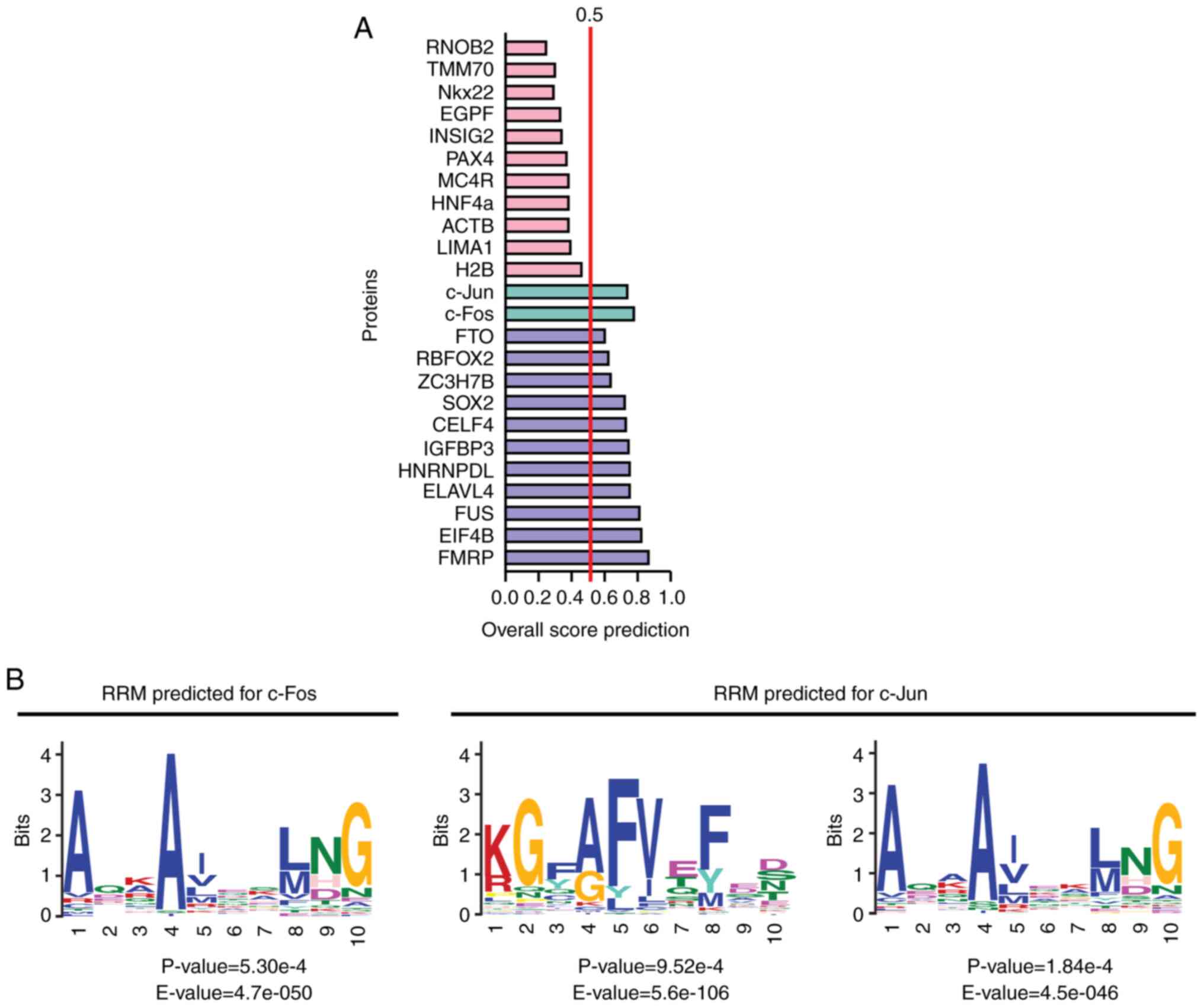
With the aim of exploring the role of direct interactions between circRNAs/proteins in the pathophysiology of DN, our previous study performed an in silico analysis of the ability of proteins involved in the AGE-RAGE pathway to bind circRNAs expressed in renal cells (https://www.uacm.edu.mx/Portals/0/adam/Content/sshLYDUxokSSYJT-rhWTqg/Text/Geceta_22.pdf, unpublished data). The results revealed that the transcription factors c-Jun and c-Fos were potentially able to bind RNA according to bioinformatics analysis conducted using the server catRAPID signature (2020, RNA System Biology Italian Institute of Technology; http://s.tartaglialab.com/page/catrapid_group), which calculates the overall RNA-binding propensity of a protein and predicts its RNA-binding regions (Fig. 3). The global interaction score was 0.72 and 0.77 for c-Jun and c-Fos respectively, which was >0.5 (the threshold recommended by the algorithm) (121) and similar to those scores of known RBPs (Fig. 3A). This finding suggested the presence of RNA-binding regions in the transcription factors c-Jun and c-Fos. According to those predictions, c-Fos would belong to the classical RNA-binding class, while c-Jun would belong to the putative one (https://www.uacm.edu.mx/Portals/0/adam/Content/sshLYDUxokSSYJT-rhWTqg/Text/Geceta_22.pdf, unpublished data).
Notably, c-Jun has two regions with RNA-binding propensity: Amino acids 65-115 and 144-215. In these regions, two potential RNA recognition motifs (RRMs) were identified by MEME Suite (https://meme-suite.org/meme/tools/meme) and Tomtom tool (https://meme-suite.org/meme/tools/tomtom) (122,123). For this analysis, 66 amino acid sequences with the binding domains of 35 RBPs were used (Table SII). These RRMs were located between amino acids 193-202 and 147-154, and were similar to the domains SH3 LIG_G3BP_FGDF_1 and LIG_NBox_RRM1, respectively (Fig. 3B) (https://www.uacm.edu.mx/Portals/0/adam/Content/sshLYDUxokSSYJT-rhWTqg/Text/Geceta_22.pdf, unpublished data). By contrast, c-Fos harbors three regions with RNA-binding propensity (amino acids 90-145, 200-257 and 264-353). Only one RRM was identified in c-Fos at amino acids 217-226, which was similar to the domain LIG_NBox_RRM1, and the one found in c-Jun. This RRM was also present in ≥50% of the amino acid sequences with the binding domains of RBPs used to analyze the potential RNA-binding regions of c-Fos and c-Jun (Fig. 3B and Table SII) (https://www.uacm.edu.mx/Portals/0/adam/Content/sshLYDUxokSSYJT-rhWTqg/Text/Geceta_22.pdf, unpublished data).
Our previous study also predicted (https://www.uacm.edu.mx/Portals/0/adam/Content/sshLYDUxokSSYJT-rhWTqg/Text/Geceta_22.pdf, unpublished data) which circRNAs reported by Memczak et al (12) that were expressed in response to 25 mM glucose in 293 cells were able to interact with c-Jun and/or c-Fos using the server RPI-seq (http://pridb.gdcb.iastate.edu/RPISeq/) (124). A total of 17 circRNAs that potentially interacted with c-Jun and/or c-Fos were predicted, of which 5 potentially interacted with c-Jun and c-Fos, 3 with c-Fos and 9 with c-Jun, according to the results of classifiers support vector machine and random forest, and simultaneous prediction score with two classifiers ≥0.8 (Table I). These circRNAs had potential binding motifs to RBP, some of which are known motifs whereas others are unknown (https://www.uacm.edu.mx/Portals/0/adam/Content/sshLYDUxokSSYJT-rhWTqg/Text/Geceta_22.pdf, unpublished data) according to the predictions of RPB suite (http://www.csbio.sjtu.edu.cn/bioinf/RBPsuite/) (125).
Table ICircRNAs expressed in the 25 mM glucose-treated 293 cells that potentially interact with c-Jun and/or c-Fos. |
These results require experimental demonstration since i) 293 cells derived from human embryonic kidney cells, and circRNA expression could be dependent on developmental stage; ii) the increase in glucose concentration could lead to different variations in circRNA expression in embryonic and adult cells (19,22,26); and iii) the expression profile of the predicted circRNAs in DN is not known. It should be noted that the original kidney cell culture that served as source of 293 cells was heterogeneous and such cells are not considered an in vitro model of typical kidney cells (126). However, the majority of predicted circRNAs that interact with c-Jun and c-Fos were expressed in normal human kidney tissues (https://www.uacm.edu.mx/Portals/0/adam/Content/sshLYDUxokSSYJT-rhWTqg/Text/Geceta_22.pdf, unpublished data) according to the results of CircAtlas 3.0 (https://ngdc.cncb.ac.cn/circatlas) (Table I). Nevertheless, it is a preliminary approach to identify potential interactions between circRNAs and c-Jun or c-Fos.
It has been proposed that circRNAs can recruit transcription factors to the promoters of target genes, and may activate or inhibit their transcription (127). Thus, the interaction of c-Jun and c-Fos with circRNAs could regulate the expression of genes under the control of these transcription factors, as well as the expression of circRNAs, their parental genes or their miRNA targets through transcription factor sequestration via a negative feedback mechanism. It has also been described that, during hyperglycemia, the expression of c-Jun and c-Fos is increased, which was shown to be mediated by the AGE-RAGE interaction, leading to the activation of the JAK2 axis, and the production of collagen and other proteins of the ECM (128).
Mesangial cells exposed to high-glucose concentrations have also been reported to activate protein kinase C, which in turn can modulate the transcription factor AP-1 (formed by the association of c-Fos and c-Jun), thus increasing both the mRNA and protein expression levels of c-Fos and c-Jun in the nucleus. This increase in AP-1 is also correlated with an increased production of ECM proteins (including FN, laminin and ColIV) (128). Thus, it may be hypothesized that circRNAs interacting with c-Jun and c-Fos could exert a similar effect on the mRNA expression levels of ECM components, whereby the potential interactions between circRNAs and these transcription factors may explain some of the events associated with the onset and progression of DN.
In addition, the aforementioned predictions revealed in our previous studies (https://www.uacm.edu.mx/Portals/0/adam/Content/sshLYDUxokSSYJT-rhWTqg/Text/Geceta_22.pdf, unpublished data) suggested that c-Jun and c-Fos may be DNA-binding proteins and RBPs which must be validated in vitro or/and in vivo, as well as the mechanisms by which these potential interactions could be involved in cell processes associated with DN.
6. CircRNAs as biomarkers of DN
The intrinsic characteristics of circRNAs, namely stability and abundance, make them promising biomarkers for diagnosing and evaluating DN progression or treatment efficacy. Although circRNA expression in renal cells may provide insights into the modulation of the pathophysiological process of DN, the release of these circRNAs into the blood or urine would facilitate their use as clinical biomarkers.
Upregulation of hsa_circRNA_0003928 and downregulation of its target miR-151-3p have been observed in the serum of patients with DN (86), but its possible application as a diagnostic biomarker has not been sufficiently explored. In addition, exosomal circRNA_DLGAP4 isolated from high glucose-treated mesangial cells, patients with DN and DN rat models promotes diabetic kidney disease progression by sponging miR-143 and targeting the receptor tyrosine-protein kinase erbB-3/NF-κB/72 kDa type IV collagenase axis (76) (Fig. 2 and Table SI). A large number of the circRNAs expressed in renal cells are carried by exosomes, which are frequently involved in the pathophysiological processes associated with DN (129). It has been reported that high-glucose concentrations cause circRNA differential expression in exosomes derived from human renal tubular epithelial cells compared with controls (129,130).
Feng et al (130) reported that 7-10% of urinary exosome transcripts correspond to circRNAs. Urinary exosomes mainly arise from every epithelial cell of the nephron, while blood exosomes are not able to pass through the glomerular membrane into the urine (131). For that reason, urinary exosomes may also be implicated in the pathophysiological process of DN and could be a robust biomarker of the disease. miRNAs enriched in urinary exosomes have been associated with the progression of DN or with the early stages of this disease (132,133). However, the diagnostic potential of urinary exosomal circRNAs in DN has not yet been determined. A recent study revealed that expression of hsa_circRNA_0036649 in urinary exosomes is associated with renal function and fibrosis degree in patients with chronic kidney disease, which is a hallmark of DN (134). Other studies that have supported the feasibility of the identification of circRNAs enriched in urinary exosomes as biomarkers for kidney diseases have focused on the dysregulation of these molecules in patients with idiopathic membranous nephropathy or with immunoglobulin A nephropathy compared with healthy controls (135,136). In summary, future studies should be conducted to explore the use of serum and urine circRNAs as biomarkers for the diagnosis and prognosis of DN.
7. Conclusions
Research related to the role of non-coding RNAs as regulators of gene expression has increased in recent years. Although some authors consider that circRNAs are modulators for the initiation and development of DN, the role of these molecules in the pathophysiological process of this disease and its progression is still not fully understood. The complete profile of circRNAs expressed in the kidney, blood, or urine and their interactions, are unknown in DN. However, accumulated evidence has suggested that circRNAs may participate in the regulation of DN-related cellular processes.
CircRNAs are highly stable and abundant molecules, which makes them important potential clinical biomarkers, therapeutic targets, or novel diagnostic agents. However, numerous questions regarding these RNA molecules and their roles in DN remain to be addressed. Therefore, further studies are required to reveal the function of circRNAs in the pathophysiological processes of DN, which may also serve as the basis for developing new diagnostic and therapeutic approaches for this disease.
Supplementary Data
Availability of data and materials
Not applicable.
Authors' contributions
LLC and MBMB conceptualized the article. EAL, YPN, MBMB and LLC performed the literature search and analysis. YPN, EAL, MBMB and LLC drafted the tables and figures. LLC wrote the first draft of the manuscript. MBMB, EAL, ATC and JVF critically revised and edited the paper. Data authentication is not applicable. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Authors' information
Maximo B. Martinez Benitez, maximo.martinez@uacm.edu. mx; ORCID 0000-0003-4351-0556. Yussel Perez Navarro, yussel.perez@estudiante.uacm.edu.mx. Elisa Azuara-Liceaga, elisa.azuara@uacm.edu.mx; ORCID 0000-0002-6392-170X. Angeles Tecalco Cruz, angeles.tecalco@uacm.edu.mx; ORCID 0000-0001-9199-3834. Jesus Valdes Flores, jvaldes@cinvestav.mx; ORCID 0000-0003-1787-9229. Lilia Lopez-Canovas, lilia.lopez.canovas@uacm.edu.mx; ORCID 0000-0003-0711-2569.
Acknowledgments
The authors would like to thank to Mr. Alfredo Padilla Barberi (Postgraduate Program in Genomic Sciences, Science and Technology School, Autonomous University of Mexico City for his help in the composition of the figures.
Funding
This work was supported by the Science and Technology School, Autonomous University of Mexico City [grant no. CCyT-2022-02].
References
|
American Diabetes Association: 2. Classification and diagnosis of diabetes: Standards of medical care in diabetes-2021. Diabetes Care. 44(Suppl 1): S15–S33. 2021. View Article : Google Scholar | |
|
Sun H, Saeedi P, Karuranga S, Pinkepank M, Ogurtsova K, Duncan BB, Stein C, Basit A, Chan JCN, Mbanya JC, et al: IDF diabetes atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res Clin Pract. 183:1091192022. View Article : Google Scholar | |
|
Harding JL, Pavkov ME, Magliano DJ, Shaw JE and Gregg EW: Global trends in diabetes complications: A review of current evidence. Diabetologia. 62:3–16. 2019. View Article : Google Scholar | |
|
Prasad RB and Groop L: Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel). 6:87–123. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Lyssenko V and Laakso M: Genetic screening for the risk of type 2 diabetes: Worthless or valuable? Diabetes Care. 36(Suppl 2): S120–S126. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Vassy JL and Meigs JB: Is Genetic testing useful to predict type 2 diabetes? Best Pract Res Clin Endocrinol Metab. 26:189–201. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Miranda-Lora AL, Vilchis-Gil J, Juárez-Comboni DB, Cruz M and Klünder-Klünder M: A genetic risk score improves the prediction of type 2 diabetes mellitus in mexican youths but has lower predictive utility compared with non-genetic factors. Front Endocrinol (Lausanne). 12:6478642021. View Article : Google Scholar : PubMed/NCBI | |
|
Willems SM, Mihaescu R, Sijbrands EJG, Van Duijn CM and Janssens AC: A methodological perspective on genetic risk prediction studies in type 2 diabetes: Recommendations for future research. Curr Diab Rep. 11:511–518. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Fava S and Hattersley AT: The role of genetic susceptibility in diabetic nephropathy: evidence from family studies. Nephrol Dial Transplant. 17:1543–1546. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Szymañski M, Barciszewska MZ, Zywicki M and Barciszewski J: Noncoding RNA transcripts. J Appl Genet. 44:1–19. 2003.PubMed/NCBI | |
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 19:141–157. 2013. View Article : Google Scholar : | |
|
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al: Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss, et al: Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Guo JU, Agarwal V, Guo H and Bartel DP: Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI | |
|
Salzman J, Chen RE, Olsen MN, Wang PL and Brown PO: Cell-Type specific features of circular RNA expression. PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Song P, Yang F, Jin H and Wang X: The regulation of protein translation and its implications for cancer. Signal Transduct Target Ther. 6:682021. View Article : Google Scholar : PubMed/NCBI | |
|
Liao W, Du J, Wang Z, Feng Q, Liao M, Liu H, Yuan K and Zeng Y: The role and mechanism of noncoding RNAs in regulation of metabolic reprogramming in hepatocellular carcinoma. Int J Cancer. 151:337–347. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Ebbesen KK, Hansen TB and Kjems J: Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017. View Article : Google Scholar : | |
|
Chen LL: The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J and Li H: The emerging landscape of circular RNA in life processes. RNA Biol. 14:992–999. 2017. View Article : Google Scholar : | |
|
Barrett SP and Salzman J: Circular RNAs: Analysis, expression and potential functions. Development. 143:1838–1847. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Liu CX and Chen LL: Circular RNAs: Characterization, cellular roles, and applications. Cell. 185:2016–2034. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Chi T, Lin J, Wang M, Zhao Y, Liao Z and Wei P: Non-Coding RNA as biomarkers for type 2 diabetes development and clinical management. Front Endocrinol (Lausanne). 12:6300322021. View Article : Google Scholar : PubMed/NCBI | |
|
Wang F and Zhang M: Circ_001209 aggravates diabetic retinal vascular dysfunction through regulating miR-15b-5p/COL12A1. J Transl Med. 19:2942021. View Article : Google Scholar : PubMed/NCBI | |
|
Fan W, Pang H, Xie Z, Huang G and Zhou Z: Circular RNAs in diabetes mellitus and its complications. Front Endocrinol (Lausanne). 13:8856502022. View Article : Google Scholar : PubMed/NCBI | |
|
Patil NS, Feng B, Su Z, Castellani CA and Chakrabarti S: Circular RNA mediated gene regulation in chronic diabetic complications. Sci Rep. 11:237662021. View Article : Google Scholar : PubMed/NCBI | |
|
Tu C, Wang L, Wei L and Jiang Z: The role of circular RNA in diabetic nephropathy. Int J Med Sci. 19:916–923. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Liu R, Zhang M and Ge Y: Circular RNA HIPK3 exacerbates diabetic nephropathy and promotes proliferation by sponging miR-185. Gene. 765:1450652021. View Article : Google Scholar | |
|
van Zonneveld AJ, Kölling M, Bijkerk R and Lorenzen JM: Circular RNAs in kidney disease and cancer. Nat Rev Nephrol. 17:814–826. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Lasda E and Parker R: Circular RNAs: Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Petkovic S and Müller S: RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 43:2454–2465. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Yamazaki T, Fujiwara N, Yukinaga H, Ebisuya M, Shiki T, Kurihara T, Kioka N, Kambe T, Nagao M, Nishida E and Masuda S: The Closely Related RNA helicases, UAP56 and URH49, Preferentially Form Distinct mRNA Export Machineries and Coordinately Regulate Mitotic Progression. Mol Biol Cell. 21:2953–2965. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Huang C, Liang D, Tatomer DC and Wilusz JE: A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 32:639–644. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Ren L, Jiang Q, Mo L, Tan L, Dong Q, Meng L, Yang N and Li G: Mechanisms of circular RNA degradation. Commun Biol. 5:13552022. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang C, Huang S, Zhuang H, Ruan S, Zhou Z, Huang K, Ji F, Ma Z, Hou B and He X: YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene. 39:4507–4518. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Chang W and Wang J: Exosomes and their noncoding RNA cargo are emerging as new modulators for diabetes mellitus. Cells. 8:8532019. View Article : Google Scholar : PubMed/NCBI | |
|
Hentze MW and Preiss T: Circular RNAs: Splicing's enigma variations. EMBO J. 32:923–925. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function as efficient microRNA sponges. Nature. 495:384–388. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, Wang G, Wu P, Wang H, Jiang L, et al: Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol Cancer. 18:1162019. View Article : Google Scholar : PubMed/NCBI | |
|
Chen XT, Li ZW, Zhao X, Li ML, Hou PF, Chu SF, Zheng JN and Bai J: Role of Circular RNA in kidney-related diseases. Front Pharmacol. 12:6158822021. View Article : Google Scholar : PubMed/NCBI | |
|
Wang H, Gao X, Yu S, Wang W, Liu G, Jiang X and Sun D: Circular RNAs regulate parental gene expression: A new direction for molecular oncology research. Front Oncol. 12:9477752022. View Article : Google Scholar : PubMed/NCBI | |
|
Wu N, Yuan Z, Du KY, Fang L, Lyu J, Zhang C, He A, Eshaghi E, Zeng K, Ma J, et al: Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 26:2758–2773. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al: Circ-ZNF609 Is a Circular RNA that Can Be translated and functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang Z, Yang T and Xiao J: Circular RNAs: Promising biomarkers for human diseases. EBioMedicine. 34:267–274. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and Kadener S: CircRNA Biogenesis competes with Pre-mRNA splicing. Mol Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P and Yang BB: Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Schneider T, Hung LH, Schreiner S, Starke S, Eckhof H, Rossbach O, Reich S, Medenbach J and Bindereif A: CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep. 6:313132016. View Article : Google Scholar : PubMed/NCBI | |
|
Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, et al: Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 7:124292016. View Article : Google Scholar : PubMed/NCBI | |
|
Das A, Sinha T, Shyamal S and Panda AC: Emerging role of circular RNA-protein interactions. Noncoding RNA. 7:482021.PubMed/NCBI | |
|
Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al: Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins. Cell. 149:1393–1406. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Yar Saglam SA, Alp E and Ilke Onen H: Circular RNAs and its biological functions in health and disease. Gene Expression and Phenotypic Traits. Chen YC and Chen SJ: IntechOpen; pp. 1–37. 2020 | |
|
Yang Q, Li F, He AT and Yang BB: Circular RNAs: Expression, localization, and therapeutic potentials. Mol Ther. 29:1683–1702. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Y, Lu T, Wang Q, Liu J and Jiao W: Circular RNAs: Crucial regulators in the human body (Review). Oncol Rep. 40:3119–3135. 2018.PubMed/NCBI | |
|
Wang M, Yu F, Wu W, Zhang Y, Chang W, Ponnusamy M, Wang K and Li P: Circular RNAs: A novel type of non-coding RNA and their potential implications in antiviral immunity. Int J Biol Sci. 13:1497–1506. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Yang L, Fu J and Zhou Y: Circular RNAs and their emerging roles in immune regulation. Front Immunol. 9:29772018. View Article : Google Scholar | |
|
Shao T, Pan YH and Xiong XD: Circular RNA: an important player with multiple facets to regulate its parental gene expression. Mol Ther Nucleic Acids. 23:369–376. 2020. View Article : Google Scholar | |
|
Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, Song W, Li X, Li L, Du Z, et al: A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 19:2182018. View Article : Google Scholar : PubMed/NCBI | |
|
Liu Y, Song J, Liu Y, Zhou Z and Wang X: Transcription activation of circ-STAT3 induced by Gli2 promotes the progression of hepatoblastoma via acting as a sponge for miR-29a/b/c-3p to upregulate STAT3/Gli2. J Exp Clin Cancer Res. 39:1012020. View Article : Google Scholar : PubMed/NCBI | |
|
Okholm TLH, Nielsen MM, Hamilton MP, Christensen LL, Vang S, Hedegaard J, Hansen TB, Kjems J, Dyrskjøt L and Pedersen JS: Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. NPJ Genom Med. 2:362017. View Article : Google Scholar : PubMed/NCBI | |
|
Selby NM and Taal MW: An updated overview of diabetic nephropathy: Diagnosis, prognosis, treatment goals and latest guidelines. Diabetes Obes Metab. 22(Suppl 1): S3–S15. 2020. View Article : Google Scholar | |
|
Gheith O, Farouk N, Nampoory N, Halim MA and Al-Otaibi T: Diabetic kidney disease: Worldwide difference of prevalence and risk factors. J Nephropharmacol. 5:49–56. 2015.eCollection 2016. | |
|
Brosius FC, Khoury CC, Buller CL and Chen S: Abnormalities in signaling pathways in diabetic nephropathy. Expert Rev Endocrinol Metab. 5:51–64. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Cooper ME: Interaction of metabolic and haemodynamic factors in mediating experimental diabetic nephropathy. Diabetologia. 44:1957–1972. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Makita Z, Radoff S, Rayfield EJ, Yang Z, Skolnik E, Delaney V, Friedman EA, Cerami A and Vlassara H: Advanced glycosylation end products in patients with diabetic nephropathy. N Engl J Med. 325:836–842. 1991. View Article : Google Scholar : PubMed/NCBI | |
|
Busch M, Franke S, Rüster C and Wolf G: Advanced glycation end-products and the kidney. Eur J Clin Invest. 40:742–755. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Ramasamy R, Yan SF and Schmidt AM: Receptor for AGE (RAGE): Signaling mechanisms in the pathogenesis of diabetes and its complications. Ann N Y Acad Sci. 1243:88–102. 2011. View Article : Google Scholar | |
|
Kay AM, Simpson CL and Stewart JA Jr: The role of AGE/RAGE signaling in diabetes-mediated vascular calcification. J Diabetes Res. 2016:68097032016. View Article : Google Scholar : PubMed/NCBI | |
|
Yun J, Ren J, Liu Y, Dai L, Song L, Ma X, Luo S and Song Y: Circ-ACTR2 aggravates the high glucose-induced cell dysfunction of human renal mesangial cells through mediating the miR-205-5p/HMGA2 axis in diabetic nephropathy. Diabetol Metab Syndr. 13:722021. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Q, Cang Z, Shen L, Peng W, Xi L, Jiang X, Ge X, Xu B and Huang S: circ_0037128/miR-17-3p/AKT3 axis promotes the development of diabetic nephropathy. Gene. 765:1450762021. View Article : Google Scholar | |
|
Feng T, Li W, Li T, Jiao W and Chen S: Circular RNA_0037128 aggravates high glucose-induced damage in HK-2 cells via regulation of microRNA-497-5p/nuclear factor of activated T cells 5 axis. Bioengineered. 12:10959–10970. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Fang R, Cao X, Zhu Y and Chen Q: Hsa_circ_0037128 aggravates high glucose-induced podocytes injury in diabetic nephropathy through mediating miR-31-5p/KLF9. Autoimmunity. 55:254–263. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Tang B, Li W, Ji TT, Li XY, Qu X, Feng L and Bai S: Circ-AKT3 inhibits the accumulation of extracellular matrix of mesangial cells in diabetic nephropathy via modulating miR-296-3p/E-cadherin signals. J Cell Mol Med. 24:8779–8788. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Hu W, Han Q, Zhao L and Wang L: Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-β1. J Cell Physiol. 234:1469–1476. 2019. View Article : Google Scholar | |
|
Mou X, Chen JW, Zhou DY, Liu K, Chen LJ, Zhou D and Hu YB: A novel identified circular RNA, circ-0000491, aggravates the extracellular matrix of diabetic nephropathy glomerular mesangial cells through suppressing miR-101b by targeting TGFβRI. Mol Med Rep. 22:3785–3794. 2020.PubMed/NCBI | |
|
Bai S, Xiong X, Tang B, Ji T, Li X, Qu X and Li W: Exosomal circ_DLGAP4 promotes diabetic kidney disease progression by sponging miR-143 and targeting ERBB3/NF-κB/MMP-2 axis. Cell Death Dis. 11:10082020. View Article : Google Scholar | |
|
Xu B, Wang Q, Li W, Xia L, Ge X, Shen L, Cang Z, Peng W, Shao K and Huang S: Circular RNA circEIF4G2 aggravates renal fibrosis in diabetic nephropathy by sponging miR-218. J Cell Mol Med. 26:1799–1805. 2022. View Article : Google Scholar | |
|
Yao T, Zha D, Hu C and Wu X: Circ_0000285 promotes podocyte injury through sponging miR-654-3p and activating MAPK6 in diabetic nephropathy. Gene. 747:1446612020. View Article : Google Scholar : PubMed/NCBI | |
|
Qiu B, Qi X and Wang J: CircTLK1 downregulation attenuates high glucose-induced human mesangial cell injury by blocking the AKT/NF-κB pathway through sponging miR-126-5p/miR-204-5p. Biochem Genet. 60:1471–1487. 2022. View Article : Google Scholar | |
|
Chen B, Li Y, Liu Y and Xu Z: circLRP6 regulates high glucose-induced proliferation, oxidative stress, ECM accumulation, and inflammation in mesangial cells. J Cell Physiol. 234:21249–21259. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Feng F, Yang J, Wang G, Huang P, Li Y and Zhou B: Circ_0068087 promotes high glucose-induced human renal tubular cell injury through regulating miR-106a-5p/ROCK2 pathway. Nephron. 147:212–222. 2023. View Article : Google Scholar | |
|
Zhuang L, Wang Z, Hu X, Yang Q, Pei X and Jin G: CircHIPK3 alleviates high glucose toxicity to human renal tubular epithelial HK-2 cells through regulation of miR-326/miR-487a-3p/SIRT1. Diabetes Metab Syndr Obes. 14:729–740. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Liu H, Wang X, Wang ZY and Li L: Circ_0080425 inhibits cell proliferation and fibrosis in diabetic nephropathy via sponging miR-24-3p and targeting fibroblast growth factor 11. J Cell Physiol. 235:4520–4529. 2020. View Article : Google Scholar | |
|
Wang W, Feng J, Zhou H and Li Q: Circ_0123996 promotes cell proliferation and fibrosis in mouse mesangial cells through sponging miR-149-5p and inducing Bach1 expression. Gene. 761:1449712020. View Article : Google Scholar | |
|
Li G, Qin Y, Qin S, Zhou X, Zhao W and Zhang D: Circ_WBSCR17 aggravates inflammatory responses and fibrosis by targeting miR-185-5p/SOX6 regulatory axis in high glucose-induced human kidney tubular cells. Life Sci. 259:1182692020. View Article : Google Scholar : PubMed/NCBI | |
|
An L, Ji D, Hu W, Wang J, Jin X, Qu Y and Zhang N: Interference of hsa_circ_0003928 alleviates high glucose-induced cell apoptosis and inflammation in HK-2 cells via mir-151-3p/anxa2. Diabetes Metab Syndr Obes. 13:3157–3168. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Liu Q, Cui Y, Ding N and Zhou C: Knockdown of circ_0003928 ameliorates high glucose-induced dysfunction of human tubular epithelial cells through the miR-506-3p/HDAC4 pathway in diabetic nephropathy. Eur J Med Res. 27:552022. View Article : Google Scholar : PubMed/NCBI | |
|
Ge X, Xi L, Wang Q, Li H, Xia L, Cang Z, Peng W and Huang S: Circular RNA Circ_0000064 promotes the proliferation and fibrosis of mesangial cells via miR-143 in diabetic nephropathy. Gene. 758:1449522020. View Article : Google Scholar : PubMed/NCBI | |
|
Sun L, Han Y, Shen C, Luo H and Wang Z: Emodin alleviates high glucose-induced oxidative stress, inflammation and extracellular matrix accumulation of mesangial cells by the circ_0000064/miR-30c-5p/Lmp7 axis. J Recept Signal Transduct Res. 42:302–312. 2022. View Article : Google Scholar | |
|
Wang H, Huang S, Hu T, Fei S and Zhang H: Circ_0000064 promotes high glucose-induced renal tubular epithelial cells injury to facilitate diabetic nephropathy progression through miR-532-3p/ROCK1 axis. BMC Endocr Disord. 22:672022. View Article : Google Scholar : PubMed/NCBI | |
|
Li J, Min Y and Zhao Q: Circ_0000064 knockdown attenuates high glucose-induced proliferation, inflammation and extracellular matrix deposition of mesangial cells through miR-424-5p-mediated WNT2B inhibition in cell models of diabetic nephropathy. Clin Exp Nephrol. 26:943–954. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Peng F, Gong W, Li S, Yin B, Zhao C, Liu W, Chen X, Luo C, Huang Q, Chen T, et al: circRNA_010383 Acts as a Sponge for miR-135a, and its downregulated expression contributes to renal fibrosis in diabetic nephropathy. Diabetes. 70:603–615. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Y, Qi Y, Ji T, Tang B, Li X, Zheng P and Bai S: Circ_LARP4 regulates high glucose-induced cell proliferation, apoptosis, and fibrosis in mouse mesangial cells. Gene. 765:1451142021. View Article : Google Scholar | |
|
Sun A, Sun N, Liang X and Hou Z: Circ-FBXW12 aggravates the development of diabetic nephropathy by binding to miR-31-5p to induce LIN28B. Diabetol Metab Syndr. 13:1412021. View Article : Google Scholar : PubMed/NCBI | |
|
Wu R, Niu Z, Ren G, Ruan L and Sun L: CircSMAD4 alleviates high glucose-induced inflammation, extracellular matrix deposition and apoptosis in mouse glomerulus mesangial cells by relieving miR-377-3p-mediated BMP7 inhibition. Diabetol Metab Syndr. 13:1372021. View Article : Google Scholar : PubMed/NCBI | |
|
Liu J, Duan P, Xu C, Xu D, Liu Y and Jiang J: CircRNA circ-ITCH improves renal inflammation and fibrosis in streptozotocin-induced diabetic mice by regulating the miR-33a-5p/SIRT6 axis. Inflamm Res. 70:835–846. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao L, Chen H, Zeng Y, Yang K, Zhang R, Li Z, Yang T and Ruan H: Circular RNA circ_0000712 regulates high glucose-induced apoptosis, inflammation, oxidative stress, and fibrosis in (DN) by targeting the miR-879-5p/SOX6 axis. Endocr J. 68:1155–1164. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhu Y, Zha F, Tang B, Ji TT, Li XY, Feng L and Bai SJ: Exosomal hsa_circ_0125310 promotes cell proliferation and fibrosis in diabetic nephropathy via sponging miR-422a and targeting the IGF1R/p38 axis. J Cell Mol Med. 26:151–162. 2022. View Article : Google Scholar | |
|
Jin J, Wang Y, Zheng D, Liang M and He Q: A Novel Identified Circular RNA, mmu_mmu_circRNA_0000309, Involves in Germacrone-Mediated Improvement of Diabetic Nephropathy Through Regulating Ferroptosis by Targeting miR-188-3p/GPX4 Signaling Axis. Antioxid Redox Signal. 36:740–759. 2022. View Article : Google Scholar | |
|
Chen S: Circ_000166/miR-296 aggravates the process of diabetic renal fibrosis by regulating the SGLT2 signaling pathway in renal tubular epithelial cells. Dis Markers. 2022:61030862022.PubMed/NCBI | |
|
Wang D, Zhang Z, Si Z and Wang L: Circ 0006282/miR-155 reduced inflammation in diabetic nephropathy via expression of SIRT1/NLRP3 signaling pathway. Food Sci Technol (Campinas). 42:e395202022. View Article : Google Scholar | |
|
Li Y, Yu W, Xiong H and Yuan F: Circ_0000181 regulates miR-667-5p/NLRC4 axis to promote pyroptosis progression in diabetic nephropathy. Sci Rep. 12:119942022. View Article : Google Scholar : PubMed/NCBI | |
|
Zhuang L, Jin G, Qiong W, Ge X and Pei X: Circular RNA COL1A2 mediates high glucose-induced oxidative stress and pyroptosis by regulating MiR-424-5p/SGK1 in diabetic nephropathy. Appl Biochem Biotechnol. 195:7652–7667. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Liu X and Wu Y: Circ_0000953 deficiency exacerbates podocyte injury and autophage through targeting mir-655/atg4b in diabetic nephropathy. Kidney Int Rep. 8:S198–S199. 2023. View Article : Google Scholar | |
|
Rashad NM, Sherif MH, El-Shal AS and Abdelsamad MAE: The expression profile of circANKRD36 and ANKRD36 as diagnostic biomarkers of chronic kidney disease in patients with type 2 diabetes mellitus. Egypt J Med Hum Genet. 22:432021. View Article : Google Scholar | |
|
Zhang K, Wan X, Khan MA, Sun X, Yi X, Wang Z, Chen K and Peng L: Peripheral Blood circRNA microarray profiling identities hsa_circ_0001831 and hsa_circ_0000867 as two novel circrna biomarkers for early type 2 diabetic nephropathy. Diabetes Metab Syndr Obes. 15:2789–2801. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Badr AM, Elkholy O, Said M, Fahim SA, El-Khatib M, Sabry D and Gaber RM: Diagnostic Significance of hsa_circ_0000146 and hsa_circ_0000072 biomarkers for diabetic kidney disease in patients with type 2 diabetes mellitus. J Med Biochem. 42:239–248. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Ling L, Tan Z, Zhang C, Gui S, Cui Y, Hu Y and Chen L: CircRNAs in exosomes from high glucose-treated glomerular endothelial cells activate mesangial cells. Am J Transl Res. 11:4667–4682. 2019.PubMed/NCBI | |
|
Liu M and Zhao J: Circular RNAs in diabetic nephropathy: Updates and perspectives. Aging Dis. 13:1365–1380. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Loganathan TS, Sulaiman SA, Abdul Murad NA, Shah SA, Abdul Gafor AH, Jamal R and Abdullah N: Interactions Among Non-Coding RNAs in Diabetic Nephropathy. Front Pharmacol. 11:1912020. View Article : Google Scholar : PubMed/NCBI | |
|
Xiong X, Liu C, Shen M, Yang Q, Zhao Q, Li X, Zhong X and Wang Z: Circular RNA expression profile in transgenic diabetic mouse kidneys. Cell Mol Biol Lett. 26:252021. View Article : Google Scholar : PubMed/NCBI | |
|
Bai YH, Wang JP, Yang M, Zeng Y and Jiang HY: SiRNA-HMGA2 weakened AGEs-induced epithelial-to-mesenchymal transition in tubular epithelial cells. Biochem Biophys Res Commun. 457:730–735. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Birchmeier W and Behrens J: Cadherin expression in carcinomas: Role in the formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta. 1198:11–26. 1994.PubMed/NCBI | |
|
Li JH, Wang W, Huang XR, Oldfield M, Schmidt AM, Cooper ME and Lan HY: Advanced glycation end products induce tubular epithelial-myofibroblast transition through the RAGE-ERK1/2 MAP kinase signaling pathway. Am J Pathol. 164:1389–1397. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Guria A, Sharma P, Natesan S and Pandi G: Circular RNAs-The road less traveled. Front Mol Biosci. 6:1462020. View Article : Google Scholar : PubMed/NCBI | |
|
Ikeda Y, Morikawa S, Nakashima M, Yoshikawa S, Taniguchi K, Sawamura H, Suga N, Tsuji A and Matsuda S: CircRNAs and RNA-Binding proteins involved in the pathogenesis of cancers or central nervous system disorders. Noncoding RNA. 9:232023.PubMed/NCBI | |
|
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, Yang W, Zhang C, Yang Q, Yee A, et al: A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 7:3842–3855. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Stoll L, Rodríguez-Trejo A, Guay C, Brozzi F, Bayazit MB, Gattesco S, Menoud V, Sobel J, Marques AC, Venø MT, et al: A circular RNA generated from an intron of the insulin gene controls insulin secretion. Nat Commun. 11:56112020. View Article : Google Scholar : PubMed/NCBI | |
|
Hou L, Wei Y, Lin Y, Wang X, Lai Y, Yin M, Chen Y, Guo X, Wu S, Zhu Y, et al: Concurrent binding to DNA and RNA facilitates the pluripotency reprogramming activity of Sox2. Nucleic Acids Res. 48:3869–3887. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang C, Han X, Yang L, Fu J, Sun C, Huang S, Xiao W, Gao Y, Liang Q, Wang X, et al: Circular RNA circPPM1F modulates M1 macrophage activation and pancreatic islet inflammation in type 1 diabetes mellitus. Theranostics. 10:10908–10924. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Livi CM, Klus P, Delli Ponti R and Tartaglia GG: CatRAPID signature: Identification of ribonucleoproteins and RNA-binding regions. Bioinformatics. 32:773–775. 2016. View Article : Google Scholar : | |
|
Bailey TL, Johnson J, Grant CE and Noble WS: The MEME Suite. Nucleic Acids Res. 43(W1): W39–W49. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Gupta S, Stamatoyannopoulos JA, Bailey TL and Noble WS: Quantifying similarity between motifs. Genome Biol. 8:R242007. View Article : Google Scholar : PubMed/NCBI | |
|
Muppirala UK, Honavar VG and Dobbs D: Predicting RNA-Protein interactions using only sequence information. BMC Bioinformatics. 12:4892011. View Article : Google Scholar : PubMed/NCBI | |
|
Pan X, Fang Y, Li X, Yang Y and Shen HB: RBPsuite: RNA-protein binding sites prediction suite based on deep learning. BMC Genomics. 21:8842020. View Article : Google Scholar : PubMed/NCBI | |
|
Lin YC, Boone M, Meuris L, Lemmens I, Van Roy N, Soete A, Reumers J, Moisse M, Plaisance S, Drmanac R, et al: Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat Commun. 5:47672014. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ and Xu RH: Circular RNA: Metabolism, functions and interactions with proteins. Mol Cancer. 19:1722020. View Article : Google Scholar : PubMed/NCBI | |
|
Kreisberg JI, Radnik RA, Ayo SH, Garoni J and Saikumar P: High glucose elevates c-fos and c-jun transcripts and proteins in mesangial cell cultures. Kidney Int. 46:105–112. 1994. View Article : Google Scholar : PubMed/NCBI | |
|
Xu YX, Pu SD, Li X, Yu ZW, Zhang YT, Tong XW, Shan YY and Gao XY: Exosomal ncRNAs: Novel therapeutic target and biomarker for diabetic complications. Pharmacol Res. 178:1061352022. View Article : Google Scholar : PubMed/NCBI | |
|
Feng S, LV L, Liu B, Zhu X and Jing J: MO619: Landscape RNA Profiling of Urinary Extracellular Vesicles in Patients with Diabetic Nephropathy. Nephrology Dialysis Transplantation. 37:2022. View Article : Google Scholar | |
|
Sinha N, Kumar V, Puri V, Nada R, Rastogi A, Jha V and Puri S: Urinary exosomes: Potential biomarkers for diabetic nephropathy. Nephrology (Carlton). 25:881–887. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Xie Y, Jia Y, Cuihua X, Hu F, Xue M and Xue Y: Urinary exosomal MicroRNA profiling in incipient type 2 diabetic kidney disease. J Diabetes Res. 2017:69789842017. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao Y, Shen A, Guo F, Song Y, Jing N, Ding X, Pan M, Zhang H, Wang J, Wu L, et al: Urinary Exosomal MiRNA-4534 as a novel diagnostic biomarker for diabetic kidney disease. Front Endocrinol (Lausanne). 11:5902020. View Article : Google Scholar : PubMed/NCBI | |
|
Cao Y, Shi Y, Yang Y, Wu Z, Peng N, Xiao J, Dou F, Xu J, Pei W, Fu C, et al: Urinary exosomes derived circRNAs as biomarkers for chronic renal fibrosis. Ann Med. 54:1966–1976. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Ma H, Xu Y, Zhang R, Guo B, Zhang S and Zhang X: Differential expression study of circular RNAs in exosomes from serum and urine in patients with idiopathic membranous nephropathy. Arch Med Sci. 15:738–753. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Luan R, Tian G, Ci X, Zheng Q, Wu L and Lu X: Differential expression analysis of urinary exosomal circular RNAs in patients with IgA nephropathy. Nephrology (Carlton). 26:432–441. 2021. View Article : Google Scholar : PubMed/NCBI |