Fungi and tumors: The role of fungi in tumorigenesis (Review)
- Authors:
- Published online on: March 27, 2024 https://doi.org/10.3892/ijo.2024.5640
- Article Number: 52
-
Copyright: © Cheng et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
1. Introduction
Fungi are microeukaryotes that inhabit different anatomic sites in the human body. More than 400 fungal species, mainly including three phyla, Ascomycota, Basidiomycota and Chytridiomycota, are associated with the human body (1). Over 100 fungal species, including 50 genera, are also found in mice (2). These fungi are less abundant in the human microbiome than other organisms, such as bacteria (3). However, emerging evidence has shown that fungi significantly influence host health and disease (3); for example, fungi are involved in the occurrence and development of tumors (4-7).
Tumors have complex ecosystems. They have their own unique microbiome, which includes bacteria, viruses and fungi. These intratumoral organisms participate in tumorigenesis and tumor development (8,9). Most studies of microbial dysbiosis in tumors, especially colorectal cancer (CRC), have focused on bacteria (10). However, sequencing technologies have also detected viruses, fungi and archaea in tumor tissues and revealed cancer type-specific microbial signatures (11). In 2022, Narunsky-Haziza et al (12) uncovered the fungal microbiome atlas of 35 types of cancer and demonstrated that fungi were also detected in all studied tumor types. Dohlman et al (13) also found tumor-related fungi in cancers of the gastrointestinal (GI) tract, lung, breast and head and neck by analyzing cancer genome data. Interestingly, different cancers exhibit cancer type-specific fungal profiles, such as Candida species, which are involved in the pathogenesis of CRC (3,13). Notably, multi-kingdom microbiota analyses have also provided biomarkers of CRC and bacterial-fungal interactions (14). These intratumoral fungi can be classified into six categories based on different anatomic sites, including the oral cavity, gut, adjacent tissue, lung, skin and blood circulation (15). Due to the significant enrichment of specific fungi in malignant tumors, the associations between fungi and human cancer have attracted increasing attention in recent years (3).
Multiple factors, such as interactions between bacteria and fungi, interactions between different fungi, and interactions between fungi and host factors, fungal genetic factors, and epigenetic factors, might be involved in the enrichment of fungi in tumor tissues and/or the conversion of commensal fungi to pathogenic fungi. Intratumoral fungi are potential therapeutic target(s) and/or diagnostic and prognostic indicators for tumors. These fungi are regulated by factors such as diet, fecal microbiota transplantation (FMT), probiotics, prebiotics and genetically engineered probiotics. In the present review, the associations between fungi and human cancer, cancer type-specific fungal profiles and the mechanisms by which fungi induce tumorigenesis were discussed. Furthermore, the factors that cause fungal enrichment in tumor tissues and/or the conversion of commensal fungi to pathogenic fungi, as well as potential therapeutic and preventive strategies based on intratumoral fungi were summarized.
2. Signatures of fungal species in tumors
Tumors are complicated ecosystems that are composed of cancer cells, immune cells, fibroblasts, endothelial cells and microbiota. The intratumoral microbiota is a novel and integral tumor component, which includes bacteria, that was recently identified in various cancer types. Poore et al (11) revealed cancer type-specific microbial signatures in tumor tissue. Indeed, each type of tumor has a distinct microbiota composition; for example, there is a particularly rich and diverse microbiome in breast cancer (16). Recent findings have further revealed the spatial and population heterogeneity of the intratumoral microbiome (17). These intratumoral microbiota can be used for multiple purposes, such as distinguishing normal tissue from cancer tissue, distinguishing metastatic cancers from non-metastatic cancers, distinguishing patients with cancer that respond to drugs from those that do not respond to drugs and distinguishing patients with a favorable prognosis from those with a bad prognosis (18).
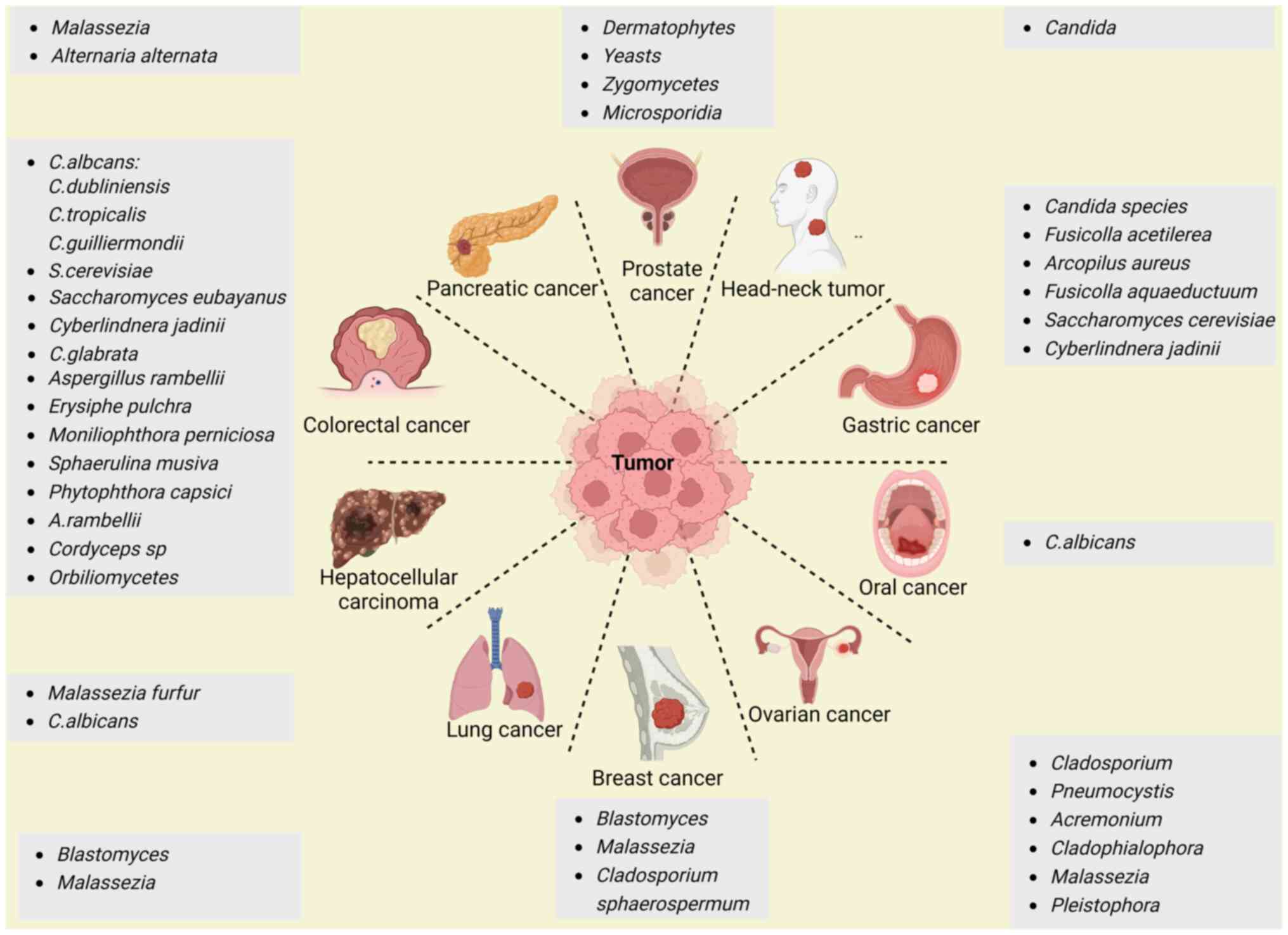
Interestingly, human tumor tissues also harbor tumor-associated fungi (12,13). For example, Narunsky-Haziza et al (12) reported that 31 fungi, such as Saccharomyces cerevisiae (99.7% coverage), were present in analyzed tumor tissues. In support of this finding, another study also revealed a high abundance and prevalence of Saccharomycetales in different tumors (13). Other fungi, including Candida albicans, Malassezia globosa, Malassezia restricta and Blastomyces gilchristii, could also be present in different types of human cancer (12,13,19). Indeed, fungi have been found in multiple types of tumors (12), such as those associated with CRC (13,20-22), pancreatic (23), breast (24), prostate (25), ovarian (26) and esophageal cancer (27). The signatures of the main specific fungi in different tumors are illustrated in Fig. 1.
CRC
CRC is the fourth most common cancer worldwide and is responsible for the deaths of >500,000 individuals every year (28). Interestingly, CRC is associated with changes in the fungal community of the colon in patients (14,21,22). Fungal dysbiosis was detected in patients with colorectal polyps (29) and adenomas (20), suggesting the involvement of fungi in early-stage CRC. Indeed, there was a co-abundance group associated with Candida albicans that included Candida dubliniensis, Candida guilliermondii and Candida tropicalis, and a group associated with Saccharomyces cerevisiae, which included Saccharomyces eubayanus, Cyberlindnera jadinii and Candida glabrata (13). These findings also indicated that GI tract cancers may be separated into Candida- and Saccharomyces-associated tumors (13). Notably, the abundance and prevalence of the species Candida dubliniensis, Candida glabrata, Candida guilliermondii, Candida lusitaniae, Candida parapsilosis, Candida tropicalis and Pichia membranifaciens were also lower in CRC according to a metagenomic analysis of whole-genome sequencing (WGS) data from multiple tumor samples from patients with different cancers in The Cancer Genome Atlas (13). Several other studies also indicated the existence of Candida species, Cyberlindnera jadinii and Saccharomyces cerevisiae in CRC tissues (30-32). However, a previous study also revealed that Aspergillus species were highly enriched in the CRC tissues of patients from both Asia and Europe through fecal shotgun metagenomic sequencing (22). In addition, other fungi, such as Cordyceps sp. RAO-2017, were also detected in CRC tissues (21). The abundance of Orbiliomycetes was different in the CRC and polyp groups (29).
Gastric cancer (GC)
GC, which is one of the most common malignancies and one of the main causes of tumor-associated deaths worldwide, is also related to fungi (33,34). A metagenomic analysis of WGS data revealed that several fungi, such as Candida species, Saccharomyces cerevisiae and Cyberlindnera jadinii, were highly abundant in the mycobiome communities of patients with GI tract cancer (13). A different study by internal transcribed spacer 2 (ITS2) analysis of GC tissues revealed significant increases in the abundance of Candida albicans, Fusicolla acetilerea, Arcopilus aureus and Fusicolla aqueductuum in cancer lesions and adjacent non-cancerous tissues of 45 patients with GC from Shenyang, China (35). Notably, the abundances of other fungi, such as Aspergillus montevidensis and Candida glabrata, were markedly reduced (35,36). Increased Candida abundance was also linked to the expression of proinflammatory factors, which could lead to the occurrence and development of tumors (13). Notably, Candida albicans might also cause GC by decreasing the diversity and richness of fungi in the stomach, which contributes to the pathogenesis of GC (35).
Hepatocellular carcinoma (HCC)
Using ITS2 rDNA sequencing, alpha diversity analyses revealed that patients with HCC had reduced fungal diversity when compared with controls (37). Aberrant colonization of the gut by Candida albicans and Malassezia furfur promoted the occurrence and development of HCC (37). HCC tumor weight and volume significantly increased in the Candida albicans and Malassezia furfur groups compared with the control group (37).
Pancreatic cancer
Pancreatic cancer, which is one of the leading causes of cancer-related deaths, is also associated with fungi. A recent preclinical and clinical study demonstrated that pancreatic ductal adenocarcinomas (PDACs) harbored significant enrichment of a specific fungus in mouse models and human specimens. Indeed, enriched fungi were observed in the pancreas of patients with PDAC and in mouse models of pancreatic cancer by principal coordinate analysis (23). Malassezia species were more prevalent in PDAC tissues in both mice and humans (23). Another analysis also demonstrated that Malassezia and Alternaria were the most abundant fungi in PDAC tumors using 18S rRNA sequencing (5). Significantly high levels of fungal and bacterial alpha diversity in the gut were also observed in patients with PDAC by 16S rRNA gene sequencing (38). Bacteria and fungi can be translocated to the pancreas and induce local and systemic changes to promote the development of PDAC (39). GFP-labeled Saccharomyces cerevisiae was detected in the pancreas of mice within 30 min of consumption (23).
Ovarian cancer
Significant differences in the abundances of Cladosporium, Pneumocystis, Acremonium, Cladophialophora, Malassezia and Pleistophora were detected in all the ovarian cancer samples. Rhizomucor, Rhodotorula, Alternaria and Geotrichum were also associated with >95% of the ovarian cancer samples according to a pan-pathogen array (PathoChip) combined with capture-next generation sequencing (26).
Prostate cancer
A fungal signature was observed in prostate cancer samples when compared with benign prostate hyperplasia samples (25). Dermatophytes (31%), yeasts (15%), Zygomycetes (15%) and Microsporidia (12%) were detected in the analyzed samples (25). The majority of fungal signatures were from the Ascomycota phylum (61%), but 50% of the fungi belonged to the class Eurotiomycetes according to hierarchical clustering analysis (25).
Breast cancer
A study revealed that Blastomyces and Malassezia species were abundant in breast tumors (13). ITS2 amplicon sequencing revealed that Cladosporium was enriched in patients with breast cancer who were ≥50 years old (12). Cladosporium was also enriched in human epidermal growth factor receptor 2-negative tumors (12). Malassezia restricta, another skin fungus, was also present in breast cancer samples (12). In addition, 7, 8 and 14% of the total hybridization signals for Ajellomyces were endocrine receptor-positive, endocrine receptor triple-positive and endocrine receptor 2-positive breast cancer, respectively, whereas Rhizomucor accounted for 19% of the hybridization signals for endocrine receptor triple-negative breast cancer (24).
Lung cancer
Blastomyces and Malassezia are associated with lung cancer (13); for instance, Blastomyces DNA was detected in 6 out of 50 patients with squamous cell lung carcinomas via metagenomic analysis of WGS data (13). Greater fungal diversity and a more complex network was also found in patients with non-small cell lung cancer (12,40).
Other tumors
A metagenomic analysis of WGS data revealed that Candida is related to head and neck tumors (13). Using Illumina™ 2×300 bp chemistry, Candida albicans was revealed to play a role in the occurrence and development of oral cancer (OC) based on the fungal ITS2 region (41).
3. Fungal-associated factors that lead to cancer
Numerous studies have shown that some specific fungi play important roles in the promotion, progression and recurrence of cancers. These fungi modulate the immune system (42), stimulate the production of specific metabolites (43,44) and potentially reconstruct different microenvironments such as biofilms. All of these factors affect not only immunity against tumors but also the genome, transcriptome, epigenome, epi-transcriptome, proteome and metabolome of tumor cells.
Immune factors
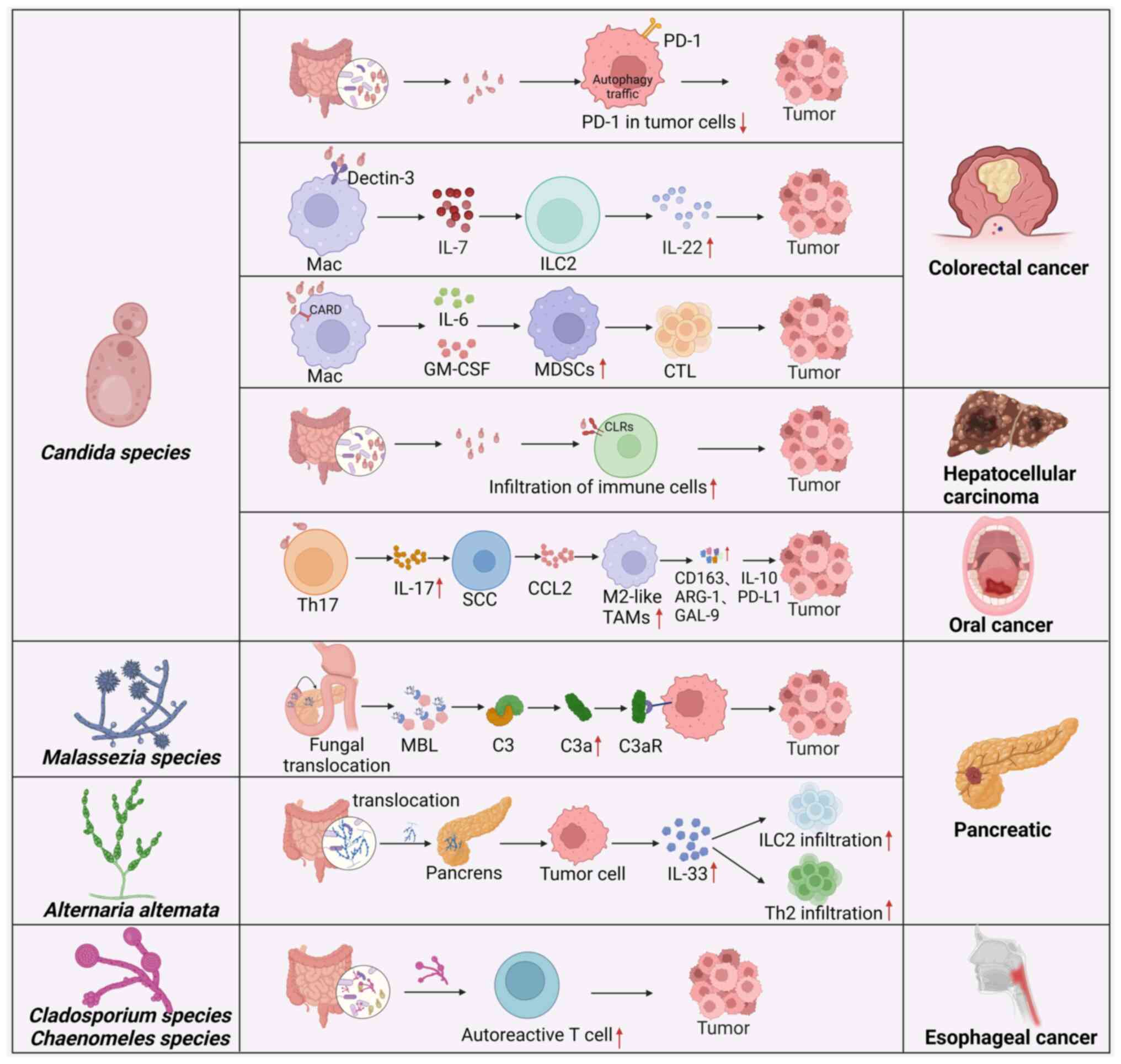
Cancers are related to fungus-mediated immune responses. Different intratumoral microbiome interactions may cause different immune responses in host tumor tissues. One study revealed three distinct clusters in tumors, termed mycotypes F1 (Malassezia-Ramularia-Trichosporon), F2 (Aspergillus-Candida) and F3 (multiple genera, including Yarrowia), which could discriminate the types of immune response, suggesting that these intratumoral mycobiomes could elicit different host responses (12). Tumors enriched with the F1 and F2 mycotypes were enriched in tumor suppressing inflammatory responses across 20 types of cancer (12). A previous study has also shown that the cell wall components of Candida guilliermondii, Candida krusei, Candida tropicalis, Candida auris and Candida albicans can trigger different types of recognition by innate immune cells in humans (42). A different study revealed the multiple mechanisms by which fungal-mediated immune factors can lead to the occurrence and development of cancers (Fig. 2). However, the aforementioned study did not examine inflammatory markers, such as C-reactive protein and albumin levels; neutrophil, lymphocyte and white blood cell counts; or the neutrophil/lymphocyte ratio, which are associated with tumor size and tissue grade in fungi-mediated tumors (45).
CRC
Myeloid-derived suppressor cells (MDSCs) are immunosuppressive cells that promote the occurrence and development of tumors. Fungal dysbiosis can increase the abundance of MDSCs, which contribute to the development of CRC. Fungal overgrowth led to the accumulation of MDSCs in the colon and worsened CRC in caspase recruitment domain 9 (CARD9)−/− mice. Treatment with the antifungal drug fluconazole suppressed CRC in CARD9−/− mice, which was associated with reduced MDSC accumulation (4). CARD9 expressed in immune cells participates in innate and adaptive immune responses via interactions between CARD9 and other molecules (46). A previous study has reported that CARD9 promotes colitis-associated cancer (47). Mutations in CARD9 are strongly associated with increased susceptibility to both fungal infections and inflammatory bowel disease in humans (48). Interestingly, when bone marrow cells were cocultured with Candida tropicalis, Candida tropicalis promoted the differentiation and function of MDSCs. In germ-free mice mono-colonized with Candida tropicalis, there was also an abundance of MDSCs in the colon (4). Further studies demonstrated that gut fungi promoted the immunosuppressive function of MDSCs by pyruvate kinase M1/2-dependent glycolysis, which promoted colorectal tumorigenesis (32). Multiple studies have reported that aerobic glycolysis is essential for MDSCs in tumors (49,50). To maintain immunosuppressive activities, MDSCs in tumors increase the level of glycolysis. Notably, MDSCs are able to absorb intratumoral glucose in the tumor microenvironment (TME) (51). However, Malik et al (6) reported that the fungal-mediated signaling axis, which is mediated by CARD9 and its upstream activator spleen tyrosine kinase (SYK), could also hinder CRC development by inducing inflammasome activation. Deletion of CARD9 or SYK in MDSCs inhibited inflammasome activation and interleukin (IL)-18 maturation and enhanced susceptibility to CRC after fungal exposure (6). Supplementation with MDSCs or IL-18 decreased the tumor burden in azoxymethane/dextran sulfate sodium (AOM/DSS)-treated CARD9−/− and SYKfl/flLysMCre/+ mice, whereas antifungal agents promoted colitis and CRC development (6).
In addition, Candida albicans can trigger glycolysis in macrophages and induce the production of IL-7, which causes the secretion of IL-22 in RAR-related orphan receptor gamma t innate lymphoid cells (ILCs) via the aryl hydrocarbon receptor and signal transducer and activator of transcription 3 to promote the progression of CRC (52). A previous study also demonstrated that the development of Candida tropicalis-mediated CRC involved reducing tumor cell-intrinsic programmed cell death protein 1 (PD-1) levels through autophagy (7). Autophagy inhibitors and Candida tropicalis treatment can limit CRC tumor growth and reverse downregulation of PD-1 expression. This finding suggested that Candida tropicalis can promote CRC progression by controlling the expression of PD-1 on tumor cells (7).
Pancreatic cancer
Analysis of PDAC revealed that Alternaria alternata, but not Candida or Aspergillus, led to the secretion of IL-33 in tumors, thereby promoting the recruitment of type 2 immune cells to promote tumor development (5). Indeed, single-cell analyses of CD45+ cells from a mouse model of pancreatic cancer revealed the presence of T helper 2 cells (TH2) and ILC2 cells, which were hallmarks of type II immune responses (5). Genetic deletion of IL-33 or antifungal treatment decreased TH2 and ILC2 infiltration and increased survival in mice. IL-33 knockdown in tumor cells in an orthotopic model demonstrated that reduced IL-33 levels decreased the infiltration of type 2 immune cells and tumor growth. Treatment with the antifungal drug amphotericin B or IL-33 depletion caused a significant decrease in tumor burden, increased survival and reduced the number of tumor-infiltrating ILC2 and TH2 cells. TH2 cells, which infiltrate the pancreas in the early stages of tumorigenesis, can produce type 2 cytokines such as IL-4 and IL-13, which promote the metabolic reprogramming of cancer cells in murine KrasG12D-driven PDAC. Consistent with type 2 immune responses that induce PDAC progression in mouse models, patients with PDAC with predominant TH2-polarized cell infiltration also exhibited reduced survival compared with patients with more TH1 cells (53). Notably, ILC2s are also present in tumors from patients with pancreatic cancer (5), and high IL-33 expression is observed in ~20% of human patients with PDAC (5).
However, the fungal community in PDAC was markedly enriched in Malassezia species in both mice and humans (23). The ligated product of mannose-binding lectin (MBL) can bind to glycans in the fungal wall to activate the complement cascade, thus causing an increase in C3a. Subsequently, C3a can bind to C3a receptor (C3aR) on the surface of tumor cells to promote tumor proliferation, motility and invasion (23). Indeed, MBL or C3 deletion in the extratumoral compartment or knockdown of the C3aR in tumor cells protected against tumor growth (23). Notably, Malassezia-mediated oncogenic progression was delayed in mice lacking MBL (23). Mice that were treated with antifungal drugs and colonized with Malassezia globosa had larger tumors. Increased levels of Malassezia were observed in human pancreatic cancer samples (23).
Esophageal cancer
Autoreactive T cells and chronic fungal infection cause esophageal carcinogenesis (27). Ikkα knock-in (IkkαKA/KA) mice develop impaired central tolerance, autoinflammation, chronic fungal infection and esophageal squamous cell carcinoma (ESCC) (27). Interestingly, during this process, autoreactive CD4+ T cells are generated, which permit fungal infection and cause tissue injury and inflammation. Antifungal treatment or the depletion of autoreactive CD4+ T cells could rescue ESCC development, whereas oral fungal administration promoted ESCC development. Thus, autoreactive T cells and chronic fungal infection promote ESCC development (27). Cladosporium cladosporioides and Chaenomeles lagenaria, which are two major fungal species that colonize the oral cavities and esophagi of IkkαKA/KA mice, might spread from the oral cavity to the esophagus. Notably, fungal infection is highly related to ESCC in non-autoimmune patients (27).
OC
Candida albicans promoted OC via IL-17A/IL-17RA and macrophages (54). IL-17A neutralization and macrophage depletion reduced the number of tumor-associated macrophages and tumor size in mice with Candida albicans infection (54). Mechanistically, Candida albicans infection promoted IL-17A production by Th17 cells. Following activation of the IL-17RA signal, tumor cells can release C-C motif chemokine ligand 2 to attract macrophages to the TME, and these macrophages exhibit an immunosuppressive phenotype with upregulated expression of IL-10, arginase-1, PD-L1 and galectin-9.
HCC
The expression of fungal recognition receptors C-type lectins (CLRs), such as dectin-1, dectin-2 and dectin-3, is downregulated in HCC. The expression of these genes is related to the clinical prognosis of patients with HCC (55). CLR-triggered immune responses might enhance the effects of immunotherapy against HCC (55). The expression of CLRs was significantly related to immune infiltration and immunotherapy efficacy in HCC.
Notably, there is still absence of evidence on the role of fungi in renal cell carcinoma (RCC). Since RCC is heavily infiltrated by T cells and myeloid cells (56), future studies should first solve whether fungi infection is related to the infiltration of T cells and myeloid cells in the occurrence and development of RCC.
Metabolites and toxins
Toxins and bioactivated amines from fungi have been linked to carcinogenesis (43,44). These factors may cause genetic, epigenetic and metabolic changes. For example, Candida albicans generates nitrosamine and metabolizes ethanol to acetaldehyde (57), which is an electrophilic and genotoxic substance that affects DNA repair, oxidative stress, DNA damage and gene mutations (58). The fungus-associated metabolite aflatoxin B1 that is produced by the Aspergillus species can induce the development of HCC via highly mutagenic DNA (59). Additionally, interactions between bacteria and fungi can also induce colorectal carcinogenesis by activating butanoate metabolism (14). Two marker genes, oraS and oraE, in the D-arginine metabolism pathway were significantly enhanced in CRC samples (14). Differential abundance analyses of the mycobiome also suggested that increased Candida abundance could promote metastasis, cellar adhesion, extracellular matrix-receptor interactions and focal adhesion (19).
Biofilms
Another possible mechanism by which the microbiota affects tumorigenesis is the formation of biofilms (60). Candida albicans can cooperate with bacteria such as E. faecalis to produce biofilms. Biofilms are closely related to CRC based on the enhancement of precancerous inflammation and escaping the host immune response (61). Interestingly, biofilm homogenates from patients with CRC can cause colon tumorigenesis in mice (62).
Fungal extracellular vesicles (EVs)
Fungal EVs can be isolated from yeast and filamentous fungi. The pathogenic role of fungal EVs has been widely reviewed (63-65). They carry pigments, carbohydrates, proteins, nucleic acids, lipids and prions, which modulate the immune responses of host cells and are tightly related to virulence (63). Furthermore, EVs play pivotal roles in orchestrating fungal communities, bolstering pathogenicity and mediating interactions with the environment (64,66). EVs from Candida albicans and Saccharomyces brasiliensis activate dendritic cells to produce cytokines such as IL-12p40, IFN-γ, TNF-α, IL-10 and TGF-β (67). EVs from pathogenic fungi also promote the production of TNF-α, TGF-β and nitric oxide by macrophages (66,68). Exophiala dermatitidis EVs could induce cell death. Understanding the function of fungal EVs can provide new and specific targets for antifungal drugs. However, there is lack of studies on the effects of fungal EVs on tumorigenesis.
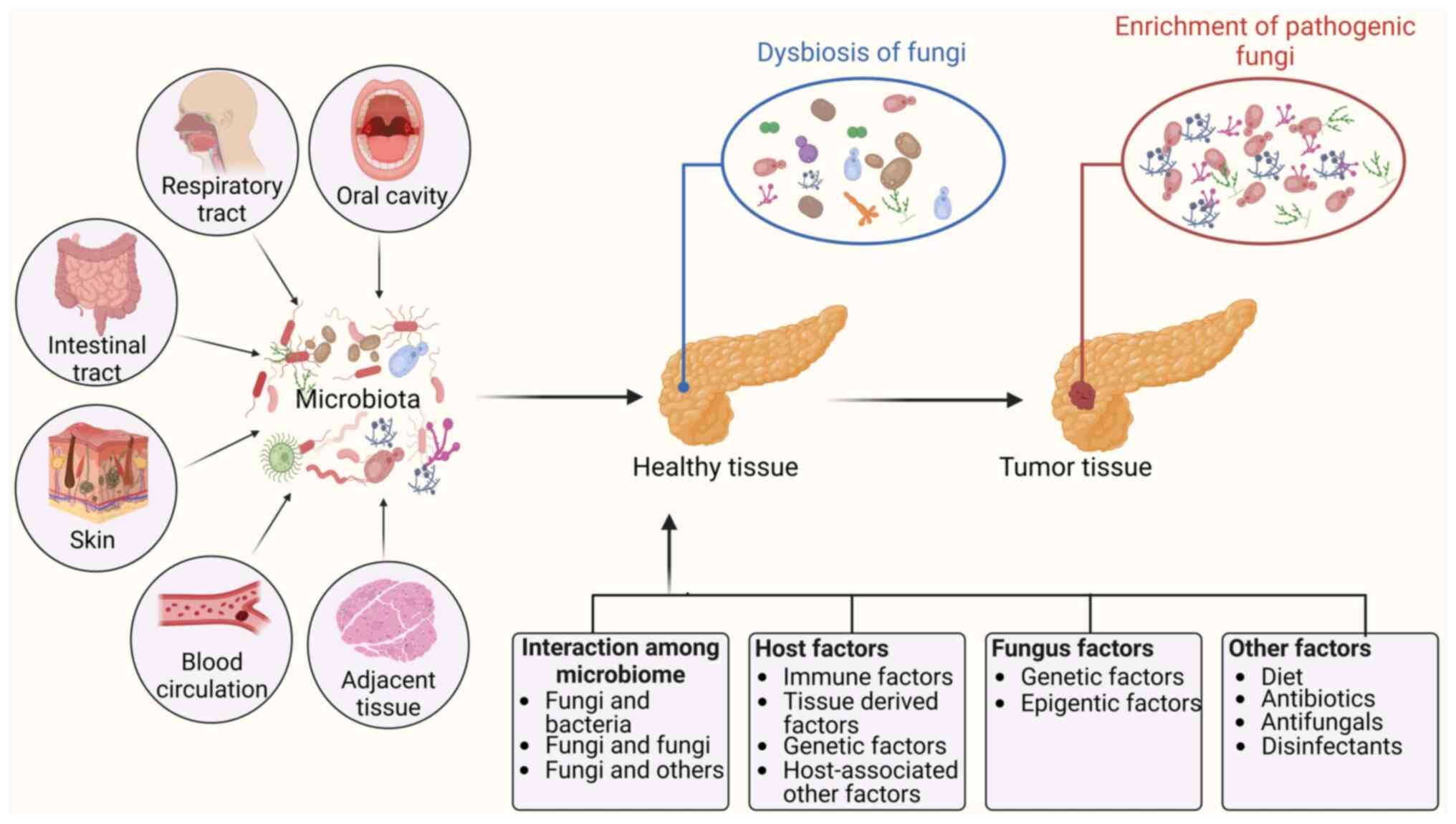
4. Factors related to the enrichment and carcinogenicity of fungal species
Intratumoral fungi can come from different anatomic sites, including the oral cavity, the gut, adjacent normal tissue, the lung, skin and blood circulation (15). Multiple factors are potentially related to the enrichment and carcinogenicity of fungal species, including interactions between microbes such as fungi and bacteria, host factors including immune factors, tissue-derived factors, and fungal genetic and epigenetic factors (Fig. 3). Notably, fungi not only are the causative agents of diseases but are also isolated from mammals without diseases (69-71), suggesting that there are two fungal types, namely, commensal and pathogenic fungi (72). Indeed, the pathogenicity of some fungi depends on their ability to change from a commensal to a pathogenic fungus (73). Li et al (74) reported that Candida albicans can aggravate intestinal inflammation by inducing proinflammatory phenotypes in vivo.
Interactions among the microbiome
Multiple different kinds of microbiota exist in the organs and tissues of humans, such as the gut. These organisms live together and form complex and dynamic ecosystems to impact host health (75). Multiple kingdom analyses of fecal samples from patients with CRC revealed strong interkingdom interactions between bacteria and fungi (14,22). A different study also revealed four kingdom microbiota alterations using metagenomic datasets from 1,368 CRC samples from 8 distinct geographic cohorts. The researchers found not only significant fungal-bacterial interactions between Aspergillus rambellii and Fusobacterium nucleatum but also significant interactions between Aspergillus rambellii and Parvimonas micra in both patients with CRC and patients with adenoma (14,22). The signature of CRC-associated fungi included 6 different enriched fungi, namely, Aspergillus rambellii, Cordyceps sp. RAO-2017, Erysiphe pulchra, Moniliophthora perniciosa, Sphaerulina musiva and Phytophthora capsici. Aspergillus rambellii is closely related to the CRC-enriched bacterium Fusobacterium nucleatum (21). Notably, experimental studies have demonstrated interactions between fungi and bacteria. For example, Lactobacillus can produce metabolites to antagonize Candida albicans growth and filamentation (76,77). Reductions in short-chain fatty acid (SCFA) levels in the murine gut were associated with an increase in Candida albicans (78). The SCFAs butyrate and propionate also inhibited the growth of the yeast Pichia kudriavzevii (79). Negative correlations between Penicillium and Faecalibacterium were found in the human gut (80). In addition, bacterium-induced immunity could also limit Candida albicans colonization of the gut lumen. Anaerobic bacteria promoted the expression of cathelicidin-related antimicrobial peptide, which can eliminate Candida albicans (78). Lactobacillus exhibits an enhanced probiotic potential following coculture with Kluyveromyces marxianus (81). Notably, bacteria-fungi interactions have revealed that bacteria can shape the immune environment that controls fungi (12). Lactobacillus kefiranofaciens and Saccharomyces cerevisiae isolated from Tibetan kefir grain alleviated AOM/DSS mediated inflammation and colorectal carcinogenesis (82). Interestingly, the presence of Candida and Saccharomyces was associated with different Fusobacterium spp. in colon cancer (13). In stomach cancer, Candida was positively associated with Dialister abundance and negatively associated with Akkermansia municiphila, Ruminococcus and Barnesiella intestinihominis abundance (13).
In addition, the fungal community also affects bacteria. Candida albicans has been shown to antagonize colonization by Escherichia and Klebsiella species. Cocolonization experiments in mice confirmed that Candida albicans could limit Klebsiella colonization in the gut (83). Lactobacillus spp., especially Lactobacillus gasseri, are frequently found in the presence of Candida and Saccharomyces (13). This observation was consistent with studies reporting that the interaction between Lactobacillus spp. and Candida influences pathogenicity (76). Candida was strongly associated with Lactobacillus in GC (13). In head and neck tumors, Candida and Saccharomyces are related to similar bacteria, such as Bifidobacterium, which support intestinal barrier function in head and neck cancers (13). Fungal dysbiosis with an increased Basidiomycota: Ascomycota ratio was observed in the feces of patients with CRC (22), implying that interactions between bacteria and fungi could contribute to colorectal carcinogenesis (22,84).
Host factors
Host factors including tissue-derived, genetic, immune and other factors can affect the enrichment and/or conversion of fungi from a commensal state to a pathogenic state. There have been several reviews on fungal immunity (85,86) and the correlations between immune responses and genetics (86). Notably, tissue-derived factors were found to affect fungi such as Candida auris, which led to the observation of subpopulations of aggregative and filamentous isolates in some clinical studies (72). Host genetic factors are also related to the transition of fungi from a commensal to a pathogenic fungus. Typically, Dectin-3−/− mice exhibited an increase in pathogenic Candida albicans (52).
Fungal genetic and epigenetic factors
Multiple fungal genetic and epigenetic factors, which are related to the enrichment and carcinogenicity of fungal species, such as ume6, which is a master regulator from yeast to hyphae Candida albicans, can suppress gut colonization by promoting the expression of the hypha-specific proinflammatory protease secreted aspartic protease 6 and the hyphal cell surface adhesion protein glutathione peroxidase-like peroxiredoxin HYR1 (87). Candida albicans in the gut causes a developmental switch of the white-opaque regulator 1 transcription factor, which leads to a commensal cell type (88). Fungi can also regulate iron uptake genes via Sef1/Sfu1, which play a role in fungal virulence and colonization (89). Candida auris also activates a stress response program via mitogen-activated protein kinase HOG1, which is necessary for virulence (90). Notably, Candida species can generate numerous more phospholipases than other fungal strains (91). In intestinal inflammation, Candida can produce candidalysin, which induces damage to cause hyphal invasion across mucosal barriers (92). Additionally, set1-mediated H3K4 methylation was required for Candida albicans virulence based on controlling reactive oxygen species levels. Candida auris also modulates genome integrity, stress responses, cell filamentation and virulence via the lncRNA DINOR (93).
Other factors
Other host factors, such as diet and age, can affect the variability of the gut mycobiota (94-96). Antibiotics, antifungals and disinfectants also affect the enrichment of fungi and/or the conversion of fungi from a commensal state to a pathogenic state. For example, antibiotics can lead to an increase in Candida in the gut, oral cavity and vagina (97,98), which facilitates invasive fungal infection through bloodstream translocation from the gut (99).
5. Application of intratumoral fungi in the diagnosis and treatment of cancers
Potential therapeutic targets
Fungi can be engineered to enhance their effects on the occurrence and development of tumors. Furthermore, intratumoral fungi can also induce innate and adaptive immune responses to prevent tumor progression (6,100). Fungi, such as Capnodiales and its genus Cladosporium, which are significantly enriched in non-responders, are also associated with immunotherapy response in patients with metastatic melanoma (12). Thus, fungi in tumor tissues might be a new potential therapeutic target in cancer therapy. At present, other microbiota, such as bacteria, have been approved by the Food and Drug Administration for the treatment of cancer (101,102).
Diagnosis and prognosis evaluation
Several studies have also reported the role of intratumoral microorganisms in diagnosis (103,104). Due to the presence of tumor type- and subtype-specific fungal profiles, intratumoral fungi have the potential to be used as diagnostic tools. However, whether fungi can be used for diagnosis has not been determined. In addition, the tumor microbiome is related to the survival rates of different patients. The presence of some intratumoral fungi may be closely related to the poor prognosis of patients with tumors. For example, in GI tumors, the presence of Candida DNA is predictive of decreased survival. Narunsky-Haziza et al (12) also suggested that fungi have prognostic and diagnostic roles in tumor tissues by comparing intratumoral fungal communities with matched bacteriomes and immunomes. The associations of fungi with clinical parameters such as the detection of early-stage cancers, overall survival in breast cancer patients and immunotherapy response in melanoma patients supported the clinical application of fungi as potential biomarkers and therapeutic targets (12).
Strategies to modulate the fungal community
Multiple therapeutic strategies targeting the microbiota (105,106), such as antifungal drugs, have been used to inhibit the oncogenic progression of PDAC. As some specific fungal species are related to the occurrence and development of tumors, antifungal chemical compounds such as terbinafine, fluconazole and itraconazole could be used for tumor therapy. The combination of an antifungal drug and chemotherapy exhibited a synergistic anticancer effect against PDAC in animal models (3). Notably, broad antibiotic application also increased the risk of cancer incidence and impaired the response to immunotherapy (107).
Specific modulation of intratumoral fungi in the clinical practice is challenging. However, the factors that regulate the gut fungal community are also potential tools for therapy against tumors.
Diet
Diet and nutrition can affect the composition of the gut microbiota and are involved in CRC onset (108). Diet-induced changes in the gut microbiome depend on whether volunteers consume a plant- or animal-based diet (109).
FMT
FMT can regulate the composition of fungi to affect tumor therapy efficacy. A high abundance of Saccharomyces and Aspergillus in donor stool was associated with effective FMT, whereas reduced FMT efficacy was related to an increase in Candida albicans in donor stool. Further study revealed that Candida was negatively correlated with total saturated fatty acids and positively correlated with carbohydrates, whereas Aspergillus was negatively correlated with the recent ingestion of SCFAs. These metabolites could directly and indirectly affect the therapeutic effectiveness of FMT against tumors.
Probiotics and prebiotics
Several functions of probiotics, such as the suppression of pathogen growth by the production of certain antimicrobial mediators (110), have been reported. Prebiotics can prevent CRC development by modifying the composition of the gut microbiota (111) and exert strong preventive effects against CRC. Notably, Saccharomyces cerevisiae plays a probiotic role in CRC by promoting cancer cell apoptosis. Saccharomyces cerevisiae reduces CRC progression by modulating the microbial structure in the mucus (31). In addition, genetically engineered microbiota could also be used as a vehicle to provide metabolic support for intratumoral T cells (112), which is essential for the proper functioning of cytotoxic T cells (113).
6. Conclusion and perspective
Omics analyses of host-microbiome interactions in human health and diseases have revealed associations between fungi and human cancer. Several cancer type-specific fungi have been identified, such as Candida species in CRC, Malassezia species in pancreatic cancer and Blastomyces species in lung and breast cancer. Importantly, some specific fungal species that lead to the occurrence and development of tumors, such as Candida species, induce CRC through the accumulation of MDSCs, and Malassezia species promote pancreatic oncogenesis by activating the complement cascade. In addition, multiple factors, such as interactions among the microbiome, are related to the enrichment of type-specific fungi in tumor tissues and/or conversion from a commensal to a pathogenic fungus. A growing body of evidence has revealed the diagnostic, prognostic and therapeutic potential of intratumoral fungi in cancer. Fungal dysbiosis in the gut can be regulated by multiple factors, such as diet, FMT, probiotics and prebiotics, which potentially affect tumor development.
However, these studies are just a start for studying intratumoral fungi, and numerous questions remain to be answered: i) What determines the abundance and composition of intratumoral fungi? Studies have shown that there is abundance of fungi in tumor tissues. Furthermore, the composition of fungi in different tumor is also different. At present, it is unclear what determines the abundance and composition of intratumoral fungi. ii) What are the origins of the intratumoral fungi? Fungi can be found not only in colorectal carcinoma but also in other tumors, such as those associated with prostate, ovarian and breast cancer. But, the origins of these intratumoral fungi are incompletely clear. iii) How do intratumoral fungi bridge cancer cells and the immune system? Fungi-mediated immune factors play important roles in tumorigenesis. It is also incompletely clear how these intratumoral fungi bridge cancer and immune cells. iv) What exact mechanism(s) do specific fungi use to induce tumorigenesis? There are different mechanisms involved in fungus-mediated tumorigenesis. An exact mechanism to induce any specific tumor needs to be investigated. v) What is the difference between commensal fungi and fungi isolated from tumor tissues, such as commensal fungi in the gut and fungi in CRC and what kind of factor(s) cause the conversion of commensal fungi to pathogenic fungi? There are two kinds of fungi, commensal fungi and fungi in the tumor tissues. At present, the difference between commensal and pathogenic fungi remains unclear. In addition, it also is unclear what kinds of factor can cause the conversion of commensal fungi to pathogenic fungi. vi) What are the functional differences between intracellular tumor-resident and extracellular tumor-resident fungi? Intracellular and extracellular fungi can be found in tumor tissues. The existence and type of differences between intracellular and extracellular fungi are unclear. In addition, the functional and physiological significance of these fungi in the TME is also unclear.
The investigation of the aforementioned questions will be decisive not only for understanding the mechanism of fungi-mediated tumor development but also for new opportunities for cancer therapy and diagnosis.
Availability of data and materials
Not applicable.
Authors' contributions
WC, FL and YG wrote the original draft and created the figures. RY conceptualized the study and contributed to the writing of the final version of the manuscript. All authors read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they no competing interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 82271779; 91842302; 81970457; 81901677 and 91629102), the Tianjin Science and Technology Commission (grant no. 18JCZDJC35300), the Ministry of Science and Technology (grant no. 2016YFC1303604), the State Key Laboratory of Medicinal Chemical Biology and the Fundamental Research Funds for the Central University, Nankai University (grant no. 63191724) and the Nankai University Tianjin Application and Basis Research (grant no. 22JCQNJC00520).
References
|
Wheeler ML, Limon JJ and Underhill DM: Immunity to commensal fungi: Detente and disease. Annu Rev Pathol. 12:359–385. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al: Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 336:1314–1317. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Okuno K, Tokunaga M, Von Hoff D, Kinugasa Y and Goel A; PDAC Biomarker Working Group: Intratumoral malasseziaglobosa levels predict survival and therapeutic response to adjuvant chemotherapy in patients with pancreatic ductal adenocarcinoma. Gastroenterology. 165:502–504 e2. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Wang T, Fan C, Yao A, Xu X, Zheng G, You Y, Jiang C, Zhao X, Hou Y, Hung MC and Lin X: The Adaptor Protein CARD9 protects against colon cancer by restricting mycobiota-mediated expansion of myeloid-derived suppressor cells. Immunity. 49:504–514 e4. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, Zhang Y, Gomez EC, Morreale B, Senchanthisai S, et al: Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell. 40:153–167 e11. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Malik A, Sharma D, Malireddi RKS, Guy CS, Chang TC, Olsen SR, Neale G, Vogel P and Kanneganti TD: SYK-CARD9 Signaling axis promotes gut fungi-mediated inflammasome activation to restrict colitis and colon cancer. Immunity. 49:515–530 e5. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Qu J, Chen Q, Bing Z, Shen S, Hou Y, Lv M and Wang T: C. tropicalis promotes CRC by down-regulating tumor cell-intrinsic PD-1 receptor via autophagy. J Cancer. 14:1794–1808. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Yang L, Li A, Wang Y and Zhang Y: Intratumoral microbiota: Roles in cancer initiation, development and therapeutic efficacy. Signal Transduct Target Ther. 8:352023. View Article : Google Scholar : PubMed/NCBI | |
|
Azevedo MM, Pina-Vaz C and Baltazar F: Microbes and Cancer: Friends or Faux? Int J Mol Sci. 21:31152020. View Article : Google Scholar : PubMed/NCBI | |
|
Shkoporov AN and Hill C: Bacteriophages of the Human Gut: The 'Known Unknown' of the Microbiome. Cell Host Microbe. 25:195–209. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, Kosciolek T, Janssen S, Metcalf J, Song SJ, et al: Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 579:567–574. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Narunsky-Haziza L, Sepich-Poore GD, Livyatan I, Asraf O, Martino C, Nejman D, Gavert N, Stajich JE, Amit G, González A, et al: Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell. 185:3789–3806 e17. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Dohlman AB, Klug J, Mesko M, Gao IH, Lipkin SM, Shen X and Iliev ID: A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell. 185:3807–3822 e12. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Liu NN, Jiao N, Tan JC, Wang Z, Wu D, Wang AJ, Chen J, Tao L, Zhou C, Fang W, et al: Multi-kingdom microbiota analyses identify bacterial-fungal interactions and biomarkers of colorectal cancer across cohorts. Nat Microbiol. 7:238–250. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Wang M, Yu F and Li P: Intratumor microbiota in cancer pathogenesis and immunity: From mechanisms of action to therapeutic opportunities. Front Immunol. 14:12690542023. View Article : Google Scholar : PubMed/NCBI | |
|
Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, et al: The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 368:973–980. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Galeano Nino JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, Futran N and Houlton J: Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. 611:810–817. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Fu A, Yao B, Dong T and Cai S: Emerging roles of intratumor microbiota in cancer metastasis. Trends Cell Biol. 33:583–593. 2023. View Article : Google Scholar | |
|
Zong Z, Zhou F and Zhang L: The fungal mycobiome: a new hallmark of cancer revealed by pan-cancer analyses. Signal Transduct Target Ther. 8:502023. View Article : Google Scholar : PubMed/NCBI | |
|
Luan C, Xie L, Yang X, Miao H, Lv N, Zhang R, Xiao X, Hu Y, Liu Y, Wu N, et al: Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Sci Rep. 5:79802015. View Article : Google Scholar : PubMed/NCBI | |
|
Lin Y, Lau HC, Liu Y, Kang X, Wang Y, Ting NL, Kwong TN, Han J, Liu W, Liu C, et al: Altered mycobiota signatures and enriched pathogenic aspergillus rambellii are associated with colorectal cancer based on multicohort fecal metagenomic analyses. Gastroenterology. 163:908–921. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, Chan FKL, Sung JJY and Yu J: Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 68:654–662. 2019. View Article : Google Scholar | |
|
Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, Shadaloey SA, Wu D, Preiss P, Verma N, et al: The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 574:264–267. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Peck KN, DeMichele AM, Alwine JC and Robertson ES: Distinct microbial signatures associated with different breast cancer types. Front Microbiol. 9:9512018. View Article : Google Scholar : PubMed/NCBI | |
|
Banerjee S, Alwine JC, Wei Z, Tian T, Shih N, Sperling C, Guzzo T, Feldman MD and Robertson ES: Microbiome signatures in prostate cancer. Carcinogenesis. 40:749–764. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Alwine JC, Coukos G and Robertson ES: The ovarian cancer oncobiome. Oncotarget. 8:36225–36245. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Zhu F, Willette-Brown J, Song NY, Lomada D, Song Y, Xue L, Gray Z, Zhao Z, Davis SR, Sun Z, et al: Autoreactive T cells and chronic fungal infection drive esophageal carcinogenesis. Cell Host Microbe. 21:478–493 e7. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Gao R, Kong C, Li H, Huang L, Qu X, Qin N and Qin H: Dysbiosis signature of mycobiota in colon polyp and colorectal cancer. Eur J Clin Microbiol Infect Dis. 36:2457–2468. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Alnuaimi AD, Wiesenfeld D, O'Brien-Simpson NM, Reynolds EC, Peng B and McCullough MJ: The development and validation of a rapid genetic method for species identification and genotyping of medically important fungal pathogens using high-resolution melting curve analysis. Mol Oral Microbiol. 29:117–130. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Li JQ, Li JL, Xie YH, Wang Y, Shen XN, Qian Y, Han JX, Chen YX and Fang JY: Saccharomyces cerevisiae may serve as a probiotic in colorectal cancer by promoting cancer cell apoptosis. J Dig Dis. 21:571–582. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang Z, Zheng Y, Chen Y, Yin Y, Chen Y, Chen Q, Hou Y, Shen S, Lv M and Wang T: Gut fungi enhances immunosuppressive function of myeloid-derived suppressor cells by activating PKM2-dependent glycolysis to promote colorectal tumorigenesis. Exp Hematol Oncol. 11:882022. View Article : Google Scholar : PubMed/NCBI | |
|
Machlowska J, Baj J, Sitarz M, Maciejewski R and Sitarz R: Gastric cancer: Epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci. 21:41022020. View Article : Google Scholar | |
|
Thrift AP and El-Serag HB: Burden of gastric cancer. Clin Gastroenterol Hepatol. 18:534–542. 2020. View Article : Google Scholar | |
|
Zhong M, Xiong Y, Zhao J, Gao Z, Ma J, Wu Z, Song Y and Hong X: Candida albicans disorder is associated with gastric carcinogenesis. Theranostics. 11:4945–4956. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Vallianou N, Kounatidis D, Christodoulatos GS, Panagopoulos F, Karampela I and Dalamaga M: Mycobiome and Cancer: What is the evidence? Cancers (Basel). 13:31492021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang L, Chen C, Chai D, Li C, Qiu Z, Kuang T, Liu L, Deng W and Wang W: Characterization of the intestinal fungal microbiome in patients with hepatocellular carcinoma. J Transl Med. 21:1262023. View Article : Google Scholar : PubMed/NCBI | |
|
Del Castillo E, Meier R, Chung M, Koestler DC, Chen T, Paster BJ, Charpentier KP, Kelsey KT, Izard J and Michaud DS: The microbiomes of pancreatic and duodenum tissue overlap and are highly subject specific but differ between pancreatic cancer and noncancer subjects. Cancer Epidemiol Biomarkers Prev. 28:370–383. 2019. View Article : Google Scholar | |
|
Vitiello GA, Cohen DJ and Miller G: Harnessing the microbiome for pancreatic cancer immunotherapy. Trends Cancer. 5:670–676. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao Y, Yi J, Xiang J, Jia W, Chen A, Chen L, Zheng L, Zhou W, Wu M, Yu Z and Tang J: Exploration of lung mycobiome in the patients with non-small-cell lung cancer. BMC Microbiol. 23:812023. View Article : Google Scholar : PubMed/NCBI | |
|
Perera M, Al-Hebshi NN, Perera I, Ipe D, Ulett GC, Speicher DJ, Chen T and Johnson NW: A dysbiotic mycobiome dominated by Candida albicans is identified within oral squamous-cell carcinomas. J Oral Microbiol. 9:13853692017. View Article : Google Scholar : PubMed/NCBI | |
|
Navarro-Arias MJ, Hernández-Chávez MJ, García-Carnero LC, Amezcua-Hernández DG, Lozoya-Pérez NE, Estrada-Mata E, Martínez-Duncker I, Franco B and Mora-Montes HM: Differential recognition of Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris by human innate immune cells. Infect Drug Resist. 12:783–794. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Chang F, Syrjänen S, Wang L and Syrjänen K: Infectious agents in the etiology of esophageal cancer. Gastroenterology. 103:1336–1348. 1992. View Article : Google Scholar : PubMed/NCBI | |
|
Yang CS: Research on esophageal cancer in China: A review. Cancer Res. 40(8 Pt 1): 2633–2644. 1980.PubMed/NCBI | |
|
Hashimoto K, Nishimura S, Shinyashiki Y, Ito T and Akagi M: Characterizing inflammatory markers in highly aggressive soft tissue sarcomas. Medicine (Baltimore). 101:e306882022. View Article : Google Scholar : PubMed/NCBI | |
|
Liu X, Jiang B, Hao H and Liu Z: CARD9 Signaling, inflammation, and diseases. Front Immunol. 13:8808792022. View Article : Google Scholar : PubMed/NCBI | |
|
Bergmann H, Roth S, Pechloff K, Kiss EA, Kuhn S, Heikenwälder M, Diefenbach A, Greten FR and Ruland J: Card9-dependent IL-1β regulates IL-22 production from group 3 innate lymphoid cells and promotes colitis-associated cancer. Eur J Immunol. 47:1342–1353. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Glocker EO, Hennigs A, Nabavi M, Schäffer AA, Woellner C, Salzer U, Pfeifer D, Veelken H, Warnatz K, Tahami F, et al: A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 361:1727–1735. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Leone RD and Powell JD: Metabolism of immune cells in cancer. Nat Rev Cancer. 20:516–531. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Deng Y, Yang J, Luo F, Qian J, Liu R, Zhang D, Yu H and Chu Y: mTOR-mediated glycolysis contributes to the enhanced suppressive function of murine tumor-infiltrating monocytic myeloid-derived suppressor cells. Cancer Immunol Immunother. 67:1355–1364. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Reinfeld BI, Madden MZ, Wolf MM, Chytil A, Bader JE, Patterson AR, Sugiura A, Cohen AS, Ali A, Do BT, et al: Cell-programmed nutrient partitioning in the tumour microenvironment. Nature. 593:282–288. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhu Y, Shi T, Lu X, Xu Z, Qu J, Zhang Z, Shi G, Shen S, Hou Y, Chen Y and Wang T: Fungal-induced glycolysis in macrophages promotes colon cancer by enhancing innate lymphoid cell secretion of IL-22. EMBO J. 40:e1053202021. View Article : Google Scholar : PubMed/NCBI | |
|
De Monte L, Reni M, Tassi E, Clavenna D, Papa I, Recalde H, Braga M, Di Carlo V, Doglioni C and Protti MP: Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J Exp Med. 208:469–478. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Wang X, Wu S, Wu W, Zhang W, Li L, Liu Q and Yan Z: Candida albicans promotes oral cancer via IL-17A/IL-17RA-Macrophage axis. mBio. 14:e00447232023. View Article : Google Scholar : PubMed/NCBI | |
|
Xia J, Ding H, Liu S, An R, Shi X, Chen M and Ren H: C-Type lectin receptors-triggered antifungal immunity may synergize with and optimize the effects of immunotherapy in hepatocellular carcinoma. J Inflamm Res. 16:19–33. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Erendor F, Sahin EO, Sanlioglu AD, Balci MK, Griffith TS and Sanlioglu S: Lentiviral gene therapy vectors encoding VIP suppressed diabetes-related inflammation and augmented pancreatic beta-cell proliferation. Gene Ther. 28:130–141. 2021. View Article : Google Scholar | |
|
Gainza-Cirauqui ML, Nieminen MT, Novak Frazer L, Aguirre-Urizar JM, Moragues MD and Rautemaa R: Production of carcinogenic acetaldehyde by Candida albicans from patients with potentially malignant oral mucosal disorders. J Oral Pathol Med. 42:243–249. 2013. View Article : Google Scholar | |
|
Smith MT, Guyton KZ, Gibbons CF, Fritz JM, Portier CJ, Rusyn I, DeMarini DM, Caldwell JC, Kavlock RJ, Lambert PF, et al: Key characteristics of carcinogens as a basis for organizing data on mechanisms of carcinogenesis. Environ Health Perspect. 124:713–721. 2016. View Article : Google Scholar : | |
|
Rushing BR and Selim MI: Aflatoxin B1: A review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem Toxicol. 124:81–100. 2019. View Article : Google Scholar | |
|
Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, Felding BH, Ivanisevic J, Cho K, Wick EC, Hechenbleikner EM, et al: Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 21:891–897. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Hold GL and Allen-Vercoe E: Gut microbial biofilm composition and organisation holds the key to CRC. Nat Rev Gastroenterol Hepatol. 16:329–330. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Tomkovich S, Dejea CM, Winglee K, Drewes JL, Chung L, Housseau F, Pope JL, Gauthier J, Sun X, Mühlbauer M, et al: Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J Clin Invest. 129:1699–1712. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Garcia-Ceron D, Bleackley MR and Anderson MA: Fungal extracellular vesicles in pathophysiology. Subcell Biochem. 97:151–177. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Freitas MS, Bonato VLD, Pessoni AM, Rodrigues ML, Casadevall A and Almeida F: Fungal extracellular vesicles as potential targets for immune interventions. mSphere. 4:e00747–19. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Rodrigues ML and Casadevall A: A two-way road: Novel roles for fungal extracellular vesicles. Mol Microbiol. 110:11–15. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Freitas MS, Bitencourt TA, Rezende CP, Martins NS, Dourado TMH, Tirapelli CR and Almeida F: Aspergillus fumigatus extracellular vesicles display increased galleria mellonella survival but partial pro-inflammatory response by macrophages. J Fungi (Basel). 9:5412023. View Article : Google Scholar : PubMed/NCBI | |
|
Vargas G, Rocha JD, Oliveira DL, Albuquerque PC, Frases S, Santos SS, Nosanchuk JD, Gomes AM, Medeiros LC, Miranda K, et al: Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell Microbiol. 17:389–407. 2015. View Article : Google Scholar | |
|
Bielska E, Sisquella MA, Aldeieg M, Birch C, O'Donoghue EJ and May RC: Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat Commun. 9:15562018. View Article : Google Scholar : PubMed/NCBI | |
|
Hamad I, Ranque S, Azhar EI, Yasir M, Jiman-Fatani AA, Tissot-Dupont H, Raoult D and Bittar F: Culturomics and amplicon-based metagenomic approaches for the study of fungal population in human gut microbiota. Sci Rep. 7:167882017. View Article : Google Scholar : PubMed/NCBI | |
|
Leong C, Schmid B, Toi MJ, Wang J, Irudayaswamy AS, Goh JPZ, Bosshard PP, Glatz M and Dawson TL Jr: Geographical and ethnic differences influence culturable commensal yeast diversity on healthy skin. Front Microbiol. 10:18912019. View Article : Google Scholar : PubMed/NCBI | |
|
Chen Y, Chen Z, Guo R, Chen N, Lu H, Huang S, Wang J and Li L: Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagn Microbiol Infect Dis. 70:492–498. 2011. View Article : Google Scholar | |
|
Proctor DM, Drummond RA, Lionakis MS and Segre JA: One population, multiple lifestyles: Commensalism and pathogenesis in the human mycobiome. Cell Host Microbe. 31:539–553. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Tsui C, Kong EF and Jabra-Rizk MA: Pathogenesis of Candida albicans biofilm. Pathog Dis. 74:ftw0182016. View Article : Google Scholar : PubMed/NCBI | |
|
Li XV, Leonardi I, Putzel GG, Semon A, Fiers WD, Kusakabe T, Lin WY, Gao IH, Doron I, Gutierrez-Guerrero A, et al: Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature. 603:672–678. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Seelbinder B, Lohinai Z, Vazquez-Uribe R, Brunke S, Chen X, Mirhakkak M, Lopez-Escalera S, Dome B, Megyesfalvi Z, Berta J, et al: Candida expansion in the gut of lung cancer patients associates with an ecological signature that supports growth under dysbiotic conditions. Nat Commun. 14:26732023. View Article : Google Scholar : PubMed/NCBI | |
|
Zeise KD, Woods RJ and Huffnagle GB: Interplay between Candida albicans and lactic acid bacteria in the gastrointestinal tract: Impact on colonization resistance, microbial carriage, opportunistic infection, and host immunity. Clin Microbiol Rev. 34:e00323202021. View Article : Google Scholar : PubMed/NCBI | |
|
MacAlpine J, Daniel-Ivad M, Liu Z, Yano J, Revie NM, Todd RT, Stogios PJ, Sanchez H, O'Meara TR, Tompkins TA, et al: A small molecule produced by Lactobacillus species blocks Candida albicans filamentation by inhibiting a DYRK1-family kinase. Nat Commun. 12:61512021. View Article : Google Scholar : PubMed/NCBI | |
|
Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X, Simms-Waldrip TR, Xie Y, Hoope LV and Koh AY: Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med. 21:808–814. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Boutin RC, Petersen C, Woodward SE, Serapio-Palacios A, Bozorgmehr T, Loo R, Chalanuchpong A, Cirstea M, Lo B, Huus KE, et al: Bacterial-fungal interactions in the neonatal gut influence asthma outcomes later in life. Elife. 10:e677402021. View Article : Google Scholar : PubMed/NCBI | |
|
Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, et al: The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. 5:1532017. View Article : Google Scholar : PubMed/NCBI | |
|
Gonzalez-Orozco BD, Kosmerl E, Jiménez-Flores R and Alvarez VB: Enhanced probiotic potential of Lactobacillus kefiranofaciens OSU-BDGOA1 through co-culture with Kluyveromyces marxianus bdgo-ym6. Front Microbiol. 14:12366342023. View Article : Google Scholar : PubMed/NCBI | |
|
Zeng X, Jia H, Shi Y, Chen K, Wang Z, Gao Z, Yuan Y and Yue T: Lactobacillus kefiranofaciens JKSP109 and Saccharomyces cerevisiae JKSP39 isolated from Tibetan kefir grain co-alleviated AOM/DSS induced inflammation and colorectal carcinogenesis. Food Funct. 13:6947–6961. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Rao C, Coyte KZ, Bainter W, Geha RS, Martin CR and Rakoff-Nahoum S: Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature. 591:633–638. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Hanahan D: Hallmarks of cancer: New dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Hoft MA, Hoving JC and Brown GD: Signaling C-Type lectin receptors in antifungal immunity. Curr Top Microbiol Immunol. 429:63–101. 2020.PubMed/NCBI | |
|
Hatinguais R, Willment JA and Brown GD: PAMPs of the fungal cell wall and mammalian PRRs. Curr Top Microbiol Immunol. 425:187–223. 2020.PubMed/NCBI | |
|
Witchley JN, Penumetcha P, Abon NV, Woolford CA, Mitchell AP and Noble SM: Candida albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection. Cell Host Microbe. 25:432–443 e6. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Pande K, Chen C and Noble SM: Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 45:1088–1091. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Chen C, Pande K, French SD, Tuch BB and Noble SM: An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe. 10:118–135. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Day AM, McNiff MM, da Silva Dantas A, Gow NAR and Quinn J: Hog1 regulates stress tolerance and virulence in the emerging fungal pathogen Candida auris. mSphere. 3:e00506–18. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Deorukhkar SC, Saini S and Mathew S: Non-albicans Candida Infection: An emerging threat. Interdiscip Perspect Infect Dis. 2014:6159582014. View Article : Google Scholar : PubMed/NCBI | |
|
Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Höfs S, Gratacap RL, Robbins J, Runglall M, et al: Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature. 532:64–68. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Gao J, Chow EWL, Wang H, Xu X, Cai C, Song Y, Wang J and Wang Y: LncRNA DINOR is a virulence factor and global regulator of stress responses in Candida auris. Nat Microbiol. 6:842–851. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Boutin RCT, Sbihi H, McLaughlin RJ, Hahn AS, Konwar KM, Loo RS, Dai D, Petersen C, Brinkman FSL, Winsor GL, et al: Composition and associations of the infant gut fungal microbiota with environmental factors and childhood allergic outcomes. mBio. 12:e03396202021. View Article : Google Scholar : PubMed/NCBI | |
|
Yamaguchi N, Sonoyama K, Kikuchi H, Nagura T, Aritsuka T and Kawabata J: Gastric colonization of Candida albicans differs in mice fed commercial and purified diets. J Nutr. 135:109–115. 2005. View Article : Google Scholar | |
|
Robbins J, Passmore GM, Abogadie FC, Reilly JM and Brown DA: Effects of KCNQ2 gene truncation on M-type Kv7 potassium currents. PLoS One. 8:e718092013. View Article : Google Scholar : PubMed/NCBI | |
|
Goncalves B, Ferreira C, Alves CT, Henriques M, Azeredo J and Silva S: Vulvovaginal candidiasis: Epidemiology, microbiology and risk factors. Crit Rev Microbiol. 42:905–927. 2016. View Article : Google Scholar | |
|
Seelbinder B, Chen J, Brunke S, Vazquez-Uribe R, Santhaman R, Meyer AC, de Oliveira Lino FS, Chan KF, Loos D, Imamovic L, et al: Antibiotics create a shift from mutualism to competition in human gut communities with a longer-lasting impact on fungi than bacteria. Microbiome. 8:1332020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhai B, Ola M, Rolling T, Tosini NL, Joshowitz S, Littmann ER, Amoretti LA, Fontana E, Wright RJ, Miranda E, et al: High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat Med. 26:59–64. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Chandra D, Selvanesan BC, Yuan Z, Libutti SK, Koba W, Beck A, Zhu K, Casadevall A, Dadachova E and Gravekamp C: 32-Phosphorus selectively delivered by listeria to pancreatic cancer demonstrates a strong therapeutic effect. Oncotarget. 8:20729–20740. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA and Knight R: The microbiome and human cancer. Science. 371:eabc45522021. View Article : Google Scholar : PubMed/NCBI | |
|
Dhankhar R, Gupta V, Kumar S, Kapoor RK and Gulati P: Microbial enzymes for deprivation of amino acid metabolism in malignant cells: Biological strategy for cancer treatment. Appl Microbiol Biotechnol. 104:2857–2869. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Halley A, Leonetti A, Gregori A, Tiseo M, Deng DM, Giovannetti E and Peters GJ: The role of the microbiome in cancer and therapy efficacy: Focus on lung cancer. Anticancer Res. 40:4807–4818. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Brandi G, Turroni S, McAllister F and Frega G: The human microbiomes in pancreatic cancer: Towards evidence-based manipulation strategies? Int J Mol Sci. 22:99142021. View Article : Google Scholar : PubMed/NCBI | |
|
Fazzino L, Anisman J, Chacón JM and Harcombe WR: Phage cocktail strategies for the suppression of a pathogen in a cross-feeding coculture. Microb Biotechnol. 13:1997–2007. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Wong CC and Yu J: Gut microbiota in colorectal cancer development and therapy. Nat Rev Clin Oncol. 20:429–452. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Elkrief A, Derosa L, Kroemer G, Zitvogel L and Routy B: The negative impact of antibiotics on outcomes in cancer patients treated with immunotherapy: A new independent prognostic factor? Ann Oncol. 30:1572–1579. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Mayne ST, Playdon MC and Rock CL: Diet nutrition, and cancer: Past present and future. Nat Rev Clin Oncol. 13:504–515. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al: Diet rapidly and reproducibly alters the human gut microbiome. Nature. 505:559–563. 2014. View Article : Google Scholar : | |
|
Roy S and Dhaneshwar S: Role of prebiotics, probiotics, and synbiotics in management of inflammatory bowel disease: Current perspectives. World J Gastroenterol. 29:2078–2100. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Clark MJ, Robien K and Slavin JL: Effect of prebiotics on biomarkers of colorectal cancer in humans: A systematic review. Nutr Rev. 70:436–443. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Canale FP, Basso C, Antonini G, Perotti M, Li N, Sokolovska A, Neumann J, James MJ, Geiger S, Jin W, et al: Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature. 598:662–666. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, Kogadeeva M, Picotti P, Meissner F, Mann M, et al: L-Arginine Modulates T cell metabolism and enhances survival and anti-tumor activity. Cell. 167:829–842 e13. 2016. View Article : Google Scholar : PubMed/NCBI |