M2 macrophage‑derived exosomes alleviate KCa3.1 channel expression in rapidly paced HL‑1 myocytes via the NF‑κB (p65)/STAT3 signaling pathway
- Authors:
- Published online on: February 8, 2024 https://doi.org/10.3892/mmr.2024.13179
- Article Number: 55
-
Copyright: © Chen et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
In recent years, exosomes have been reported as a promising cell-free therapeutic strategy in inflammatory disease to assuage inflammation, pyroptosis and autophagy (1). In particular, exosomes derived from cardiomyocytes could alleviate the sensitivity of tumor ferroptosis in ischemic heart failure (2). Moreover, exosomes have been demonstrated to mitigate arrhythmogenesis and inflammasomes in arrhythmogenic cardiomyopathy (3). As far as the current study is concerned, inflammation also drives electrical remodeling and structural remodeling, ultimately leading to the perpetuation of atrial fibrillation (AF) (4). However, it is unknown whether exosomes affect AF and the specific mechanism.
Exosomes are single-membrane vesicles with a diameter of 30~200 nm. Almost all cells can release exosomes to facilitate intercellular communication and preserve proper function (5). In recent years, exosomes have been demonstrated to encapsulate critical molecules, such as DNA, mRNA, microRNA (miRNA or miR) and proteins, in response to the microenvironment of heart diseases and mediate transmission between cells (6). Numerous studies have reported that exosomes are secreted from epicardial fat and atrial myocytes, and several of them have been identified to enhance macrophage polarization, cardiac fibroblast activation and the transformation of proarrhythmic molecules in AF (7). In addition, exosomes are also important messengers that facilitate communication between macrophages and cardiomyocytes (8). In a previous study by the authors, it was found that blocking exosome release with GW4869 suppressed AF in a canine model, which was probably related to the enrichment of profibrotic miR-21-5p in exosomes (9). In another study by the authors, it was established that KCa3.1 inhibition suppressed AF susceptibility by attenuating macrophage proinflammatory polarization and inflammatory cytokine secretion in a canine model with prolonged rapid pacing (10). A recent study confirmed that M2c macrophages could reduce electrical remodeling in stem cell-derived cardiomyocytes and thereby prevent AF (11). However, to the best of the authors' knowledge, the effect of M2-exos on KCa3.1 in AF has not been evaluated. Hence, the purpose of the present study was to test the hypothesis that M2-exos have an important role in KCa3.1 expression in electrical remodeling in pacing HL-1 cells.
Materials and methods
Induction and identification of M2 macrophages
The present study was approved ((approval no. WDRM 20191211) by The Animal Research Subcommittee of the Renmin Hospital of Wuhan University Institutional Review Board, and complied with the standards established by the National Institutes of Health for the Care and Use of Laboratory Animals.
A total of 20 (10 males and 10 females; 8–12 weeks old; weight, 28–30 g) C57BL/6J mice in the present study were given free access to food and water under a clean, comfortable and 12/12-h dark/light cycle circumstance with 23~25°C, 50~60% relative humidity and noise below 60 dB. All mice were kept for 7 days after purchased, and they were healthy without any treatment after monitoring their health status indicators every other day until euthanized by cervical dislocation. None of the mice succumbed during the experiment. The criterion of verified death was the cessation of respiration, heartbeat and arterial pulsation of mice. The bone marrow macrophages were isolated from their femoral and tibial bones after euthanasia. The detailed progress is as follows.
The parietal peritoneum of mouse was opened by tissue scissors along the ventrimeson and groin as soon as possible, and attention was given to avoid damaging the visceral peritoneum. Afterward, the femoral and tibial bones were then opened with scissors and flushed with phosphate-buffered saline (PBS; containing 1% penicillin streptomycin) as soon as possible. The muscle tissue attached to the bones was carefully kicked away with tissue scissors. Next, the bones were flushed with PBS until the bone marrow color changed from red to white. Subsequently, the wash medium was centrifuged at 1,500 × g for 15 min at 4°C to discard the supernatant before adding ACK lysis buffer (cat. no. NH4CL2009; Tbdscience) and PBS. After centrifugation at 1,500 × g for 15 min at 4°C, bone marrow cells were sedimented and resuspended in high glucose medium containing 20% fetal bovine serum (Biological Industries). The cells were seeded at a density of 2×106 cells/ml, differentiated into bone marrow-derived macrophages (BMDMs) with 25 ng/ml mouse M-CSF (cat. no. 96-315-02-10; PeproTech, Inc.) for 6 days, and then cultured in complete medium with IL-4 (042149; PeproTech) at 20 ng/ml for 24 h to polarize into M2 macrophages. Next, double immunofluorescence staining and reverse transcription-quantitative PCR (RT-qPCR) were performed to identify M2 macrophages.
Isolation and authentication of exosomes
Exosomes were isolated from M2 macrophage and BMDM culture medium by ultracentrifugation. The detailed processes were carried out as described in the guidelines (12). First, the medium was collected and centrifuged at 300 × g for 15 min at 4°C to remove dead cells, and then the cell debris was removed at 3,000 × g for 15 min at 4°C. Next, the supernatant was centrifuged at 10,000 × g for 30 min at 4°C and concentrated with an ultrafiltration apparatus at 1,500 × g for 30 min at 4°C. After the concentrated liquid was centrifuged for 90 min at 120,000 × g, exosomes were extracted and then resuspended in PBS. Nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM) were performed to identify exosomes, and the exosome-specific proteins CD81 and TSG101 were quantified using western blotting.
AF model establishment at the cellular level
HL-1 myocytes were purchased from Procell Life Science & Technology Co., Ltd. and cultured in DMEM/F12 with 100 U/ml penicillin, 100 µg/ml streptomycin, and 10% fetal bovine serum (Biological Industries). HL-1 myocytes were randomly divided into six groups: Control group, pacing group, pacing + coculture group (pacing HL-1 cells cocultured with M2-exos), pacing + mimic-miR-146a-5p group, pacing + NC-miR-146a-5p group and pacing + pyrrolidine dithiocarbamate (PDTC). In addition, the miR-146a-5p mimics (mimic group) and native control (NC group) were transfected into the HL-1 myocytes in order to make sure the mimics transfection is successful. The protocol of AF model at HL-1 cells is described below. The cells were used at a density of −60–80% confluence. The sterilized carbon electrodes were placed into the plates to contact the medium. The cells were stimulated rapidly for 72 h at 10 Hz, 1.0 V/cm [stimulation system (Master 8, Israel)].
Coculture of M2-exos and HL-1 atrial myocytes
The PKH26 Red Fluorescent Cell Linker Mini Kit (cat. no. MX4021; MK BIO Science, Co., Inc.) was used to stain the M2-exos, DAPI (10 mg/ml; cat. no. D8417-1-MG; Sigma-Aldrich; Merck KGaA) was used to stain HL-1 myocyte nuclei, and the cytoskeleton was visualized using phalloidin (cat. no. 40735ES75; Shanghai Yeasen Biotechnology Co., Ltd.). All procedures were carried out in accordance with the manufacturers' instructions. The protein content of the exosomes was measured using a BCA protein assay kit according to the instructions. M2-exos (60 µg/ml) were cultured with HL-1 myocytes for 48 h and observed under a fluorescence microscope after 3 h.
Mimic-miR-146a-5p transfection
To explore the effect of miR-146a-5p derived-from M2-exos on tachypacing HL-1 myocytes, mimic-miR-146a-5p were transfected into HL-1 myocytes. Mimics-miR-146a-5p and a negative control were designed and synthesized by Shanghai GenePharma Co., Ltd. The sequences of the mimic-miR-146a-5p were as follows: 5′-UGAGAACUGAAUUCCAUGGGUU-3′ (sense) and 5′CCCAUGGAAUUCAGUUCUCAUU-3′ (antisense). For the miR-146a-5p mimic control, the sequences were 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (anti-sense). In 6-well plates, BMDM-IL-4 cells were cultivated to 60–80% confluence. For 5 min, serum-free F12/DMEM was mixed separately with the mimics (30 µM) and Lipofectamine 6000 (cat. no. C0526; Beyotime Institute of Biotechnology) at room temperature. Next, the mimics and Lipofectamine were gently mixed together and incubated at room temperature for 10 min. Finally, the diluted mimics/Lipofectamine combination was introduced to the plates and incubated for 5 h before being replaced with normal cell culture media.
Whole-cell patch-clamp
Whole-cell patch-clamp was performed with a HEKA EPC-9 amplifier (HEKA Instruments, Inc.). To record the action potentials, the bath solution contained (in mM): 140 NaCl, 5.4 KCl, 1.8 CaCl2, 0.5 MgCl2, 0.33 NaHPO4, 5 HEPES, 10 D-Glucose and pH was adjusted to 7.4 with NaOH. The pipette solution contained the following (in mM): 140 K-aspartate, 5 MgCl2, 4 K2ATP, 5 HEPES and pH was adjusted to 7.2 with KOH. To record KCa3.1 currents, the external solution contained (in mM): 140 NaCl, 4.5 KCl, 2 CaCl2, 1 MgCl2, 10 D-Glucose and 10 HEPES, pH being adjusted to 7.4 with NaOH. The internal pipette solution contained (in mM): 100 K-aspartate, 40 KCl, 1 MgCl2, 5 EGTA, 10 HEPES and 4.4 CaCl2 (1 µM free Ca2+), pH being adjusted to 7.2 with KOH. Under these solution conditions, the pipettes were made of capillary tubing (Sutter Instruments) and had resistances of 3~5 MΩ. All measurements were taken at room temperature. A single cell was clamped at a holding potential of −70 mV, and the current density was measured using a stimulus voltage pattern consisting of a 100 msec test pulse from −100 to 100 mV, followed by a 1-sec test interval at the holding potential of −70 mV. The current before TRAM-34 (cat. no. HY-13519; MedChemExpress) application was digitally subtracted from the current after TRAM-34 application to obtain KCa3.1, which was the TRAM-34-sensitive current.
RNA extraction and RT-qPCR
Total RNA was extracted using TRIzol Plus (Takara Bio, Inc.) according to the manufacturer's protocol. RT-qPCR was performed as previously described (13). The expression of miR-146a-5p, CD206 and arginase was determined using the SYBR Green Master Mix kit (cat. no. G3330-100; Wuhan Servicebio Technology Co., Ltd.) according to the manufacturer's protocol. GAPDH or U6 snRNA was used as the internal control for mRNA or miRNA quantification, and the miR-146a-5p RT-qPCR thermocycling conditions were as follows: Denaturation (1 cycle) for 10 min at 95°C; annealing (40 cycles) for 15 sec at 95°C; and extension for 1 min at 60°C. Thermocycling conditions for other target genes were as follows: Predenaturation (1 cycle) for 30 sec at 90°C; denaturation (40 cycles) for 5 sec at 95°C; and annealing/extension (40 cycles) for 30 sec at 60°C. The fold-change was calculated using the 2−ΔΔCq method (14). The sequences were as follows: miR-146a-5p forward, 5′-CGCGTGAGAACTGAATTCCAT-3′ and reverse, 5′-AGTGCAGGGTCCGAGGTATT-3′; CD206 forward, 5′-GGAGGCTGATTACGAGCAGT-3′ and reverse, 5′-TCCAGGTGAACCCCTCTGAA-3′; arginase forward, 5′-AGCACTGAGGAAAGCTGGTC-3′ and reverse, 5′-TACGTCTCGCAAGCCAATGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACAT-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; and GAPDH forward, 5′-GGGTCCCAGCTTAGGTTCATC-3′ and reverse, 5′-TACGGCCAAATCCGTTCACA-3′.
Western blotting
Cell and exosome proteins were extracted on ice using RIPA lysis buffer (cat. no. CR2203104; Beyotime Institute of Biotechnology) and quantified by the BCA method (Pierce; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The proteins were denatured with loading buffer at 100°C for 10 min. Next, the proteins (30 µg per lane) were separated with 10% SDS-polyacrylamide gel electrophoresis and then transferred to 0.45-µm nitrocellulose membranes (Pierce; Thermo Fisher Scientific, Inc.) using a semidry system. The membranes were blocked with QuickBlock (cat. no. ps108; Epizyme, Inc.) at 4°C for 1 h and incubated with primary antibodies against TSG101 (1:1,000; cat. no. GB11618; Wuhan Servicebio Technology Co., Ltd.), CD81 (1:1,000; cat. no. 66866-1-Ig; Proteintech Group, Inc.), KCa3.1 (1:1,000; cat. no. 60276-1-Ig; Proteintech Group, Inc.), STAT3 (1:1,000; cat. no. 10253-2-AP; Proteintech Group, Inc.), phosphorylated (p-)STAT3 (1:1,000; cat. no. RLP0250; ImmunoWay Biotechnology), NF-κB (p65) (1:1,000; cat. no. 10745-1-AP; Proteintech Group, Inc.), p-NF-κB (p65) (1:1,000; cat. no. GB113882; Wuhan Servicebio Technology Co., Ltd.), IL-1β (1:1,000; cat. no. AF5103; Affinity Biosciences) and GAPDH (1:1,000; cat. no. GB12002; Wuhan Servicebio Technology Co., Ltd.) at 4°C overnight. Then, the membranes were incubated at room temperature for 2 h with horseradish peroxidase-conjugated secondary antibody (1:3,000; cat. no. pr30012; Proteintech Group, Inc.) to visualize the signal with the ChemiDOC™ XRS+ imaging system (BIO-RAD). GAPDH was used as an internal standard. ImageJ bundled with Java 8 software was used for densitometric analysis (National Institutes of Health).
Immunofluorescence double staining
Immunofluorescence double staining was performed to authenticate M2 macrophages and measure the expression of KCa3.1 and NF-κB (p-p65) in each group, as previously described (15). The primary antibodies used were F4/80 (1:100; cat. no. 28463-1-Ap; Proteintech Group, Inc.), CD206 (1:75; cat. no. GB113497; Wuhan Servicebio Technology Co., Ltd.), KCa3.1 (1:100; cat. no. bs-6675r; BIOSS) and NF-κB (p-p65; 1:100; cat. no. cy6367; Shanghai Abways Biotechnology Co., Ltd.).
Statistical analysis
Descriptive data are presented as the mean ± SEM. To compare two groups, an unpaired Student's t test was employed. Comparisons of more than two groups were carried out using one-way ANOVA with post hoc Tukey's multiple comparisons test. Nominal statistical significance was set at P<0.05. All data were analyzed with Graph Pad Prism9.0 (Dotmatics).
Results
Polarization of M2 macrophages and identification of exosomes
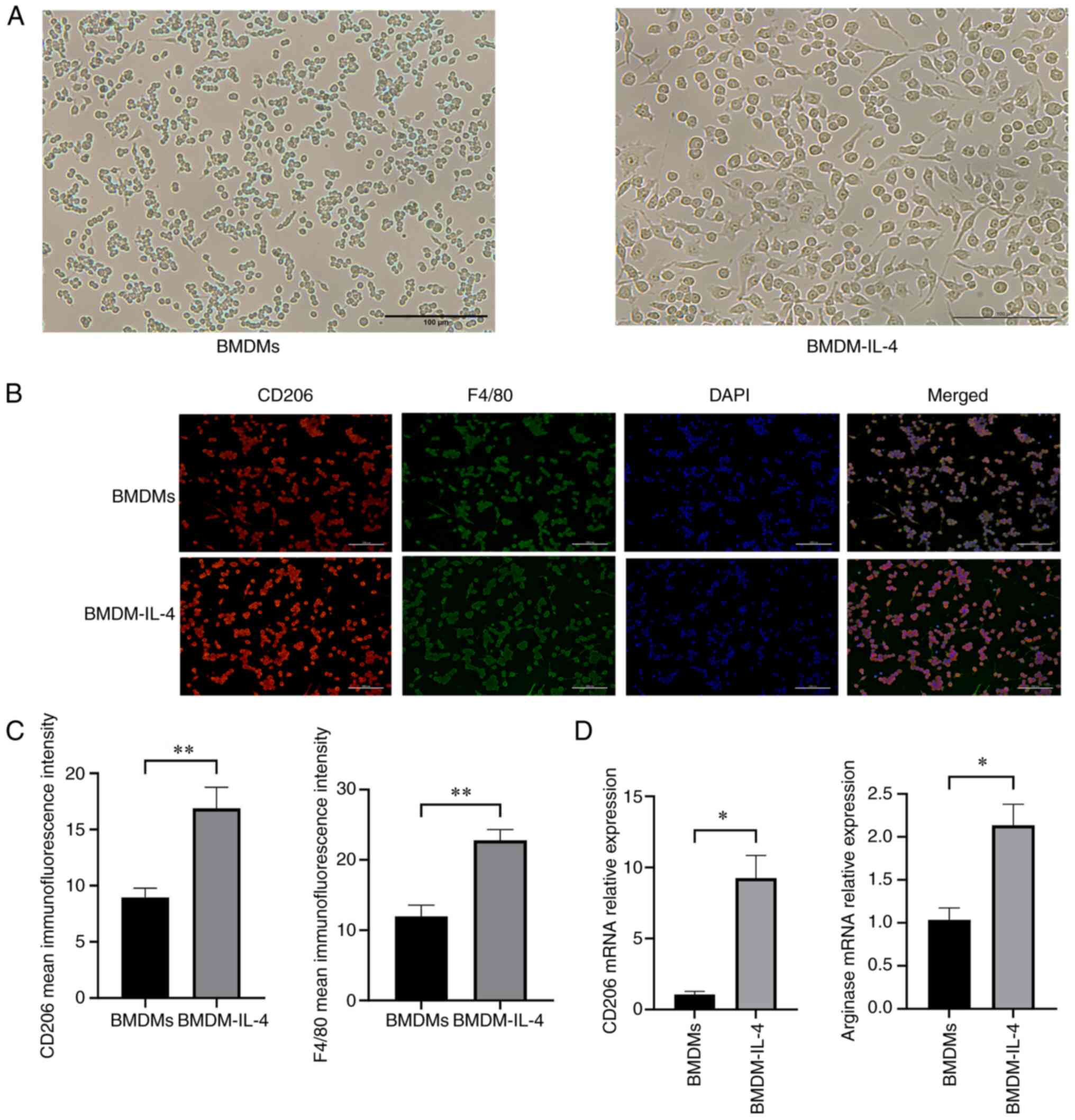
As revealed in Fig. 1A, IL-4 stimulation changed the size and morphology of BMDMs. More protrusions were observed in BMDMs in the presence of IL-4. Immunofluorescence analysis of the M2 macrophage-specific markers CD206 and F4/80 were used to identify these cells. It was obvious that the immunofluorescence densities of CD206 and F4/80 were upregulated in the BMDMs that were administrated IL-4 (P<0.01; Fig. 1B and C). At a concentration of 20 ng/ml, IL-4 increased the mRNA expression levels of markers typically linked to M2 macrophages, including arginase and CD206 (P<0.05; Fig. 1D). The results indicated that BMDMs were effectively polarized into M2 macrophages, which could be used for exosome separation.
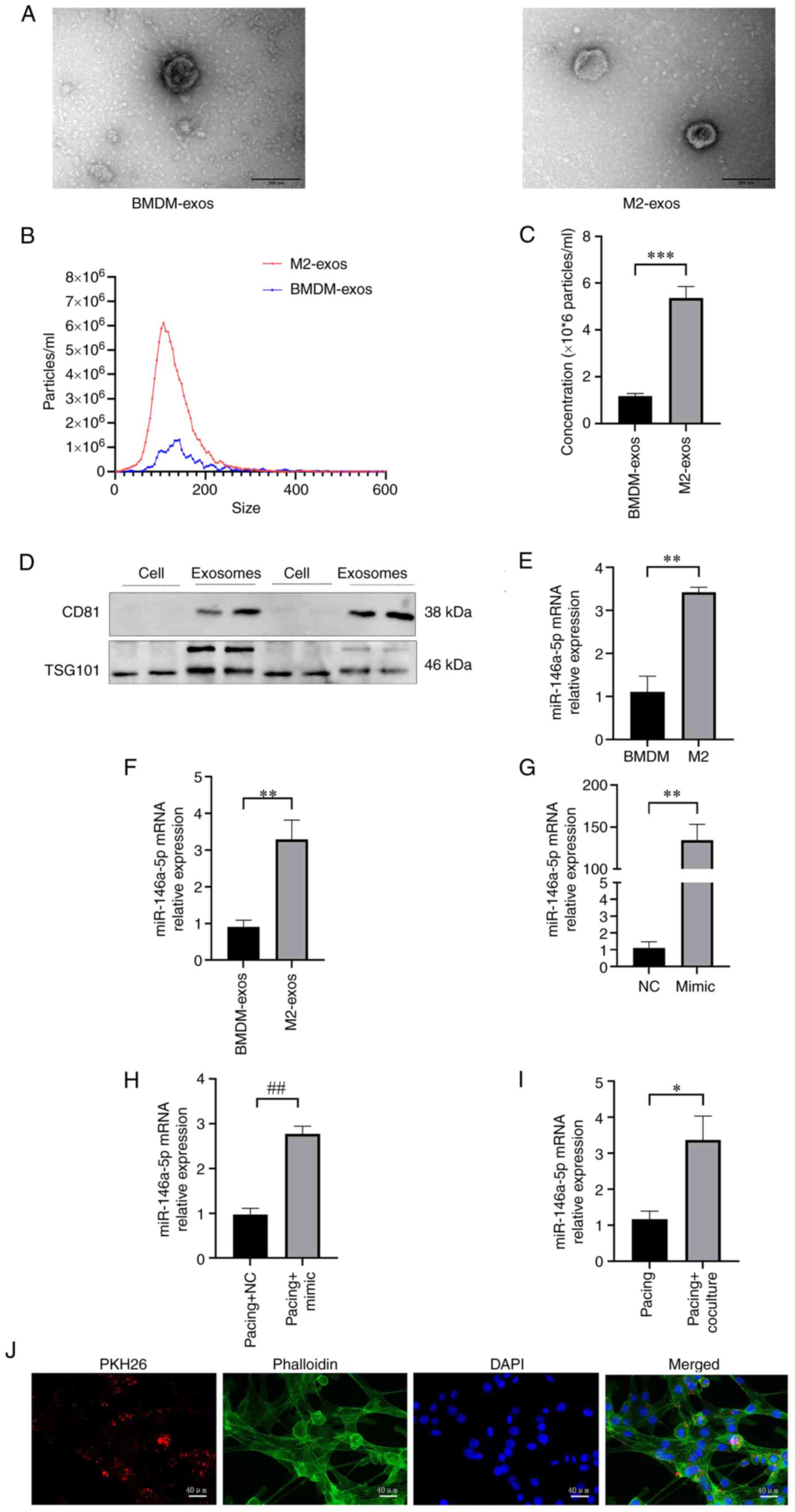
Then, exosomes from BMDMs and M2 macrophage supernatants were extracted. As demonstrated in Fig. 2A, TEM was used to examine the morphologies of exosomes. There was a little difference in the morphology between BMDM-exos and M2-exos. Numerous small particles accumulated on the surface of M2-exos, while BMDM-exos had a smoother appearance. Exosomes were also examined using NTA. In contrast with BMEM-exos, which had a mean diameter of 132.70±3.15 nm and a particle concentration of (1.17±0.12)x106/ml (P<0.001), M2-exos had a greater concentration of (5.37±0.49)x106/ml and a smaller size of 119.30±2.86 nm (P<0.01; Fig. 2B and C). The exosome-specific proteins CD81 and TSG101 were evaluated using western blotting to further confirm the purity of exosomes. Exosome proteins CD81 and TSG101 were found to be more abundant than cell proteins (Fig. 2D). Taken together, the results suggested that exosomes from M2 macrophages could be successfully isolated and used in subsequent experiments.
M2-exos carry miR-146a-5p into HL-1 myocytes
As revealed in Fig. 2E, miR-146a-5p was significantly elevated with the activation of IL-4 in M2 macrophages compared with BMDMs (P<0.01). Similarly, in contrast with BMDM-exos, miR-146a-5p levels were higher in M2-exos (P<0.01; Fig. 2F).
The miR-146a-5p mRNA relative expression in the mimic group was significantly higher than the NC group (P<0.05; Fig. 2G). Similarly, the expression of miR-146a-5p mRNA in the pacing + mimic group was significantly increased compared with the pacing + NC group (P<0.05; Fig. 2H). Then, pacing HL-1 myocytes were cocultured with M2-exos for 48 h to further investigate the role of M2-exos. The mRNA level of miR-146a-5p increased in the pacing + coculture group compared with the pacing group (P<0.05; Fig. 2I). As shown in Fig. 2J, the HL-1 myocyte cytoskeleton was tagged with phalloidin and M2-exos were labeled with PKH26. M2-exos were phagocytosed by HL-1 myocytes after coculture.
M2-exos and miR-146a-5p regulate the KCa3.1 channel in pacing HL-1 myocytes
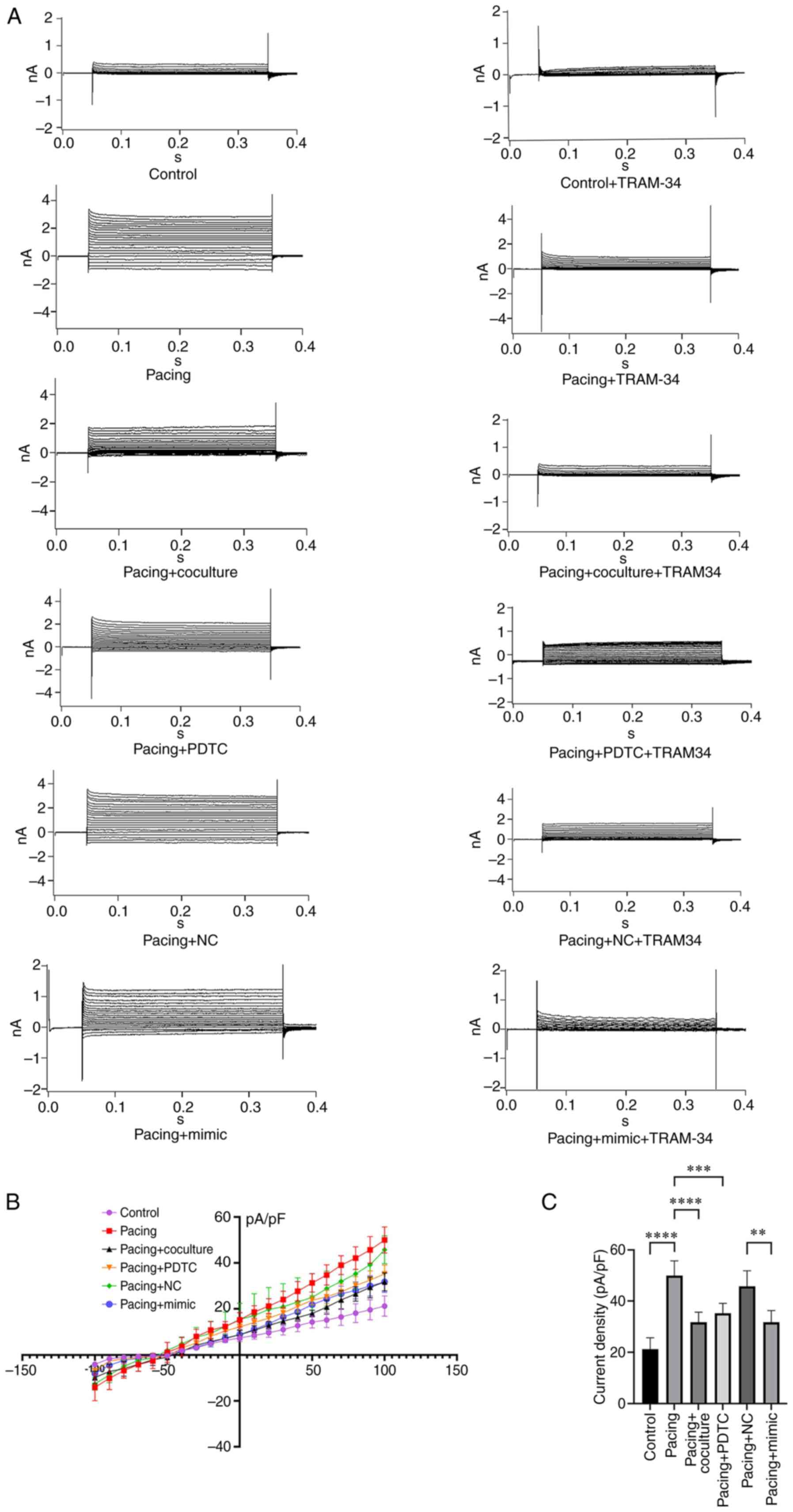
HL-1 myocyte membrane current was recorded using a whole-cell patch clamp with 300-millisec voltage steps from a holding potential of −70 mV to between −100 and +100 mV (inset). As revealed in Fig. 3A-C, the current density of KCa3.1 in pacing HL-1 myocytes was more than 2-fold higher than that in control cells (49.99±2.55 pA/pF vs. 21.23±1.98 pA/pF; n=5; P<0.0001). Pacing HL-1 myocytes treated with M2-exos had a reduced density of 31.74±1.76 pA/pF (P<0.001, Fig. 3B and C) compared with the pacing group. In addition, transporting mimic-miR-146a-5p into the pacing HL-1 myocytes led to a smaller KCa3.1 current density compared with the pacing + NC group (31.74±2.05 pA/pF vs. 45.73±2.72 pA/pF; n=5; P<0.01; Fig. 3B and C).
M2-exos and miR-146a-5p influence the action potential duration (APD) of HL-1 myocytes
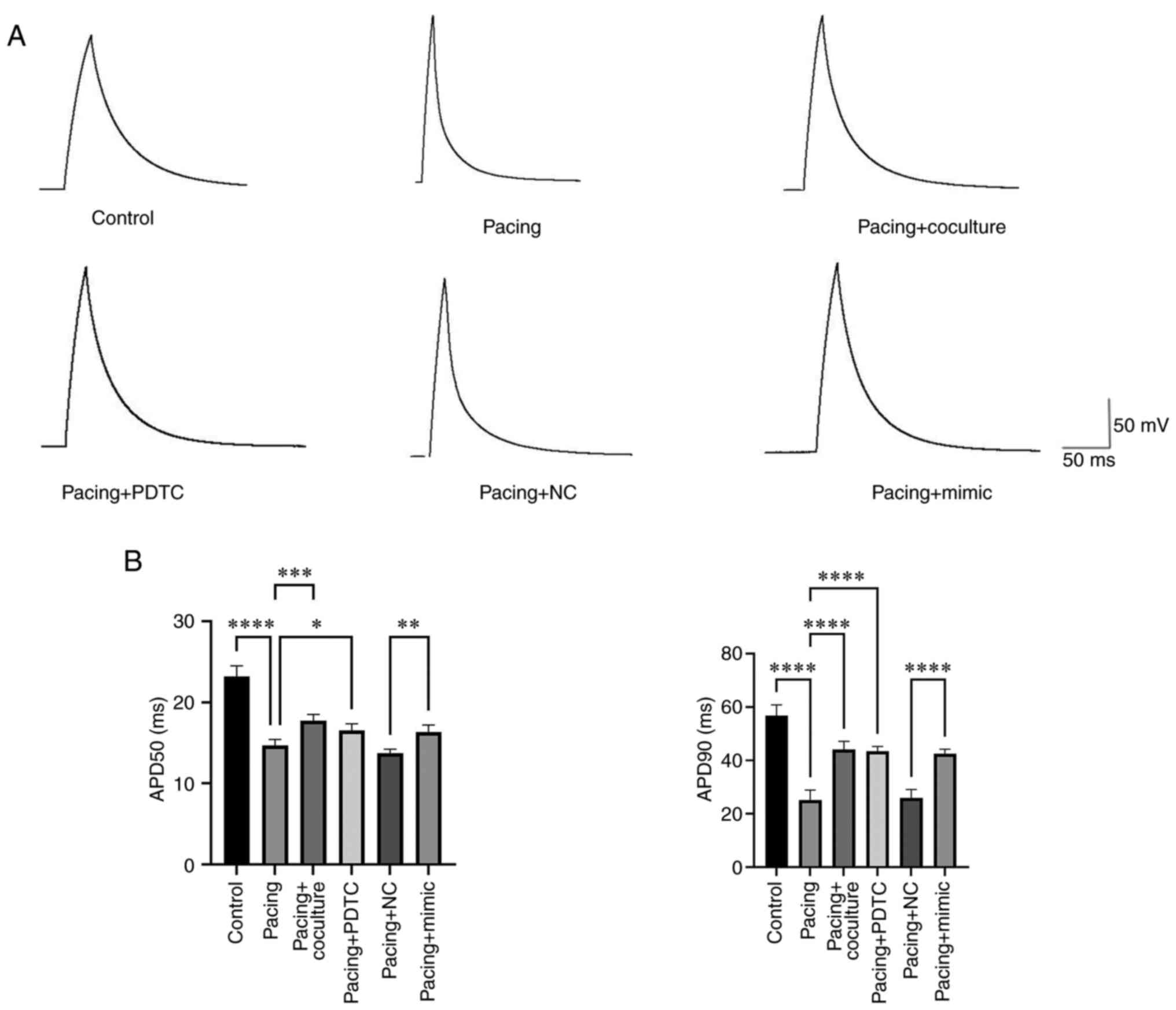
The results indicated that there was a significant reduction in APD50 and APD90 in the pacing group compared with the control group (23.18±0.59 msec vs. 14.68±0.34 msec, P<0.0001; 56.74±1.84 msec vs. 25.16±1.67 msec, P<0.0001; Fig. 4A and B). In pacing HL-1 myocytes administered M2-exos and PDTC, the APD50 and APD90 showed substantial increases compared with the pacing group (14.68±0.34 msec vs. 17.74±0.34 msec; P<0.001; 14.68±0.34 msec vs. 16.54±0.36 msec, P<0.05; 25.16±1.67 msec vs. 44.18±1.33 msec, P<0.0001; 25.16±1.67 msec vs. 43.40±0.83 msec; P<0.0001). In contrast to the Pacing + NC group, the Pacing + mimic group exhibited increases in the APD50 and APD 90 (13.76±0.21 msec vs. 16.34±0.39 msec, P<0.001; 25.88±1.43 msec vs. 42.56±0.75 msec, P<0.0001).
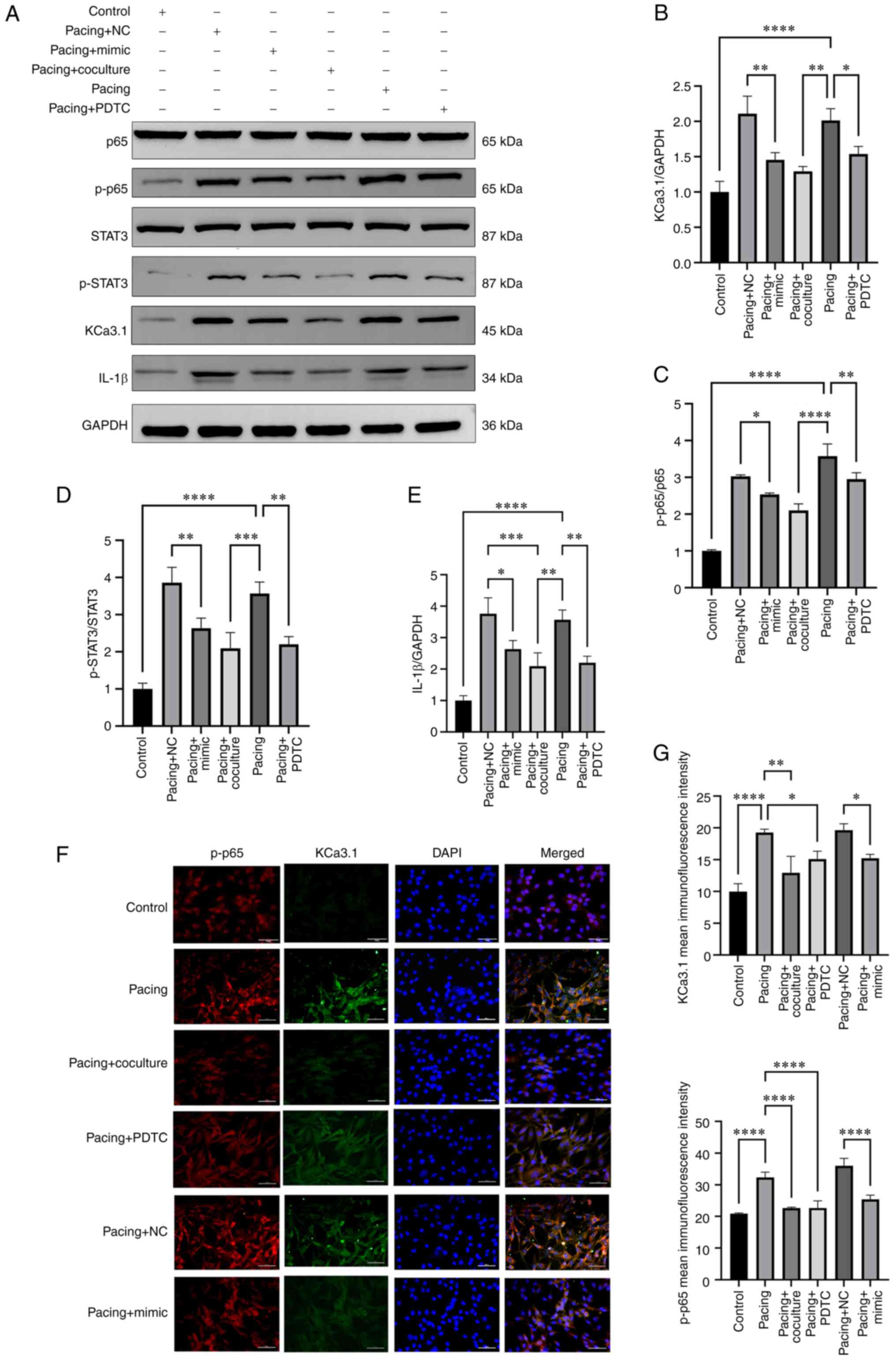
M2-exos and miR-146a-5p regulate the Ca3.1 channel via NF-κB/STAT3 in tachypacing HL-1 myocytes
As demonstrated in Fig. 5A-E, there were higher protein levels of KCa3.1, p-NF-κB p65, p-STAT3 and IL-1β in the pacing group than in the control group (P<0.0001). Pacing HL-1 cells administrated M2-exos and PDTC had significantly reduced protein levels of KCa3.1 (P<0.05), p-NF-κB p65 (P<0.01), p-STAT3 (P<0.01) and IL-1β (P<0.01) compared with the pacing group (Fig. 5A-E). In contrast to the pacing + NC group, the pacing + mimic group had the same result (P<0.05 and P<0.01). Finally, the outcomes of double immunofluorescence staining of KCa3.1 and p-NF-κB p65 revealed a similar effect (Fig. 5F and G).
Discussion
The present study, to the best of the authors' knowledge, was the first to investigate the effects of M2 macrophage-derived exosomes on KCa3.1 channels and inflammation in pacing HL-1 myocytes. The following new evidence was presented: (i) Pacing HL-1 myocytes have a higher expression of KCa3.1 current density and a shorter APD; (ii) The expression of miR-146a-5p was enriched in both M2 macrophages and M2-exos; and (iii) M2-exos and mimic-miR-146a-5p alleviated the expression of KCa3.1 current density and IL-1β secretion by suppressing the NF-κB/STAT3 signaling pathway, further prolonging the shorter APD induced by pacing HL-1 cells.
Rapid atrial stimulation can lead to atrial electrical remodeling. Studies have reported that after rapid pacing of HL-1 cells, the transcription levels of heat shock protein Hsp70, atrial natriuretic peptide and brain natriuretic peptide are significantly increased, which is highly consistent with the changes in transcription levels of human permanent AF transcriptomics such as metabolic transcription and fibrosis-related factors in vivo. Rapid pacing of HL-1 cells provided an experimental way to study AF in vitro (16). The study identified that the rapid pacing of HL-1 cells could simulate the microenvironment of AF by pacing HL-1 cardiomyocytes at 5 Hz with a 5-ms duration square wave (40 V). In the aforementioned study, HL-1 cells were inoculated into a petri dish equipped with stimulation electrodes at stimulation frequencies of 200, 300, 400, 500 and 600 bpm, and the voltage parameter was 0.8 V/cm. The current parameter was 20 mA/cm2 stimulated for 24 h, and the results indicated that under 300 bpm conditions, the duration of the cell action potential could be shortened, ICaL and ICaT ion channels could be downregulated, and outward rectified K+ channel could be upregulated, promoting the electrical remodeling of atrial cells (17). Brundel et al (18) inoculated HL-1 cells in a petri dish and gave them electric field stimulation at 10 HZ, 5.1 V/cm. Their research results revealed that rapid pacing of HL-1 cells increased the cell beating frequency by 5 times compared with the basic value, which was comparable to the degree of myocardial cell stimulation in human AF. In addition, it was found that rapid pacing significantly downregulated the expression level of L-type calcium ion channels in cells, resulting in structural changes such as myolysis and deformation and a 14-fold increase in calmodulin kinase activity. These changes were similar to myocardial cell remodeling in patients with AF. In the present study, rapid electrical stimulation of HL-1 cells was used to simulate the AF microenvironment and further improve the cell stimulation regimen.
KCa3.1 is a calcium-activated K+ channel that opens exclusively in response to increased cytosolic calcium and has a localized and functional distribution within the heart (19). Inhibition of KCa3.1 with TRAM-34 could reduce the automaticity of human embryonic stem cell-derived cardiomyocytes (20,21) and decrease delayed afterdepolarizations and arrhythmic Ca2+ transients in human induced pluripotent stem cell-derived cardiomyocytes (22). A previous study by the authors demonstrated that the expression of KCa3.1 in the left and right atrium was increased considerably after atrial rapid pacing and intravenous injection of TRAM-34 7 h later completely blocked the inducibility and duration of AF (23). Furthermore, inhibiting KCa3.1 could reduce the proinflammatory polarization of macrophages in the atrium, preventing AF in sustained fast atrial pacing (10). In the present study, the expression of KCa3.1 was further investigated at the cellular level. Pacing HL-1 cells enhanced the expression of KCa3.1 protein and increased the KCa3.1 current density. Furthermore, the APD90 and APD50 were dramatically reduced in HL-1 myocytes following fast pacing. These findings are consistent with those of our previous study (23). However, the mechanism by which KCa3.1 is regulated by rapid pacing is not clear.
MiRNAs are the major molecules within exosomes and are widely studied in cardiovascular disease (24). Previous studies have reported that upregulating miR-146a-5p expression in cardiosphere-derived cells inhibits the production of proinflammatory cytokines and transcripts, hence reducing myocardial fibrosis (25). Additionally, miR-146a-5p carried by exosomes was reported to downregulate matrix metalloprotease 2/16 expression in cardiomyocytes, which could improve cardiac contractile function and decrease susceptibility after myocardial infarction (26). Furthermore, a decreased miR-146a-5p level was linked to an increased risk of non-stroke severe adverse cardiovascular events in AF (27). In the present study, the results suggested that both M2 macrophages and M2-exos demonstrated high levels of miR-146a-5p expression, as was predicted. Pacing HL-1 myocytes that were cocultured with M2-exos had an increased expression of miR-146a-5p. Pacing HL-1 myocytes revealed shorter APD, increased expression of KCa3.1 protein and current density in their cytomembranes. Pacing HL-1 myocytes treated with M2-exos and mimic-miR-146a-5p reduced KCa3.1 protein and current density, further reversing the shorter APD caused by rapid pacing. However, the mechanism by which M2-exos and their cargo carrying miR-146a-5p cargo affect pacing cells is unknown.
Subsequently, the particular mechanism through which miR-146a-5p regulates the KCa3.1 channel was investigated. First, the miR-146a-5p mimics were transfected into the HL-1 myocytes. Obviously, the miR-146a-5p mRNA had a higher expression in the mimic group than the NC group. The result demonstrated that the mimic transfection was successful. It has been reported that the NF-κB-dependent gene miR-146a-5p may directly inhibit the production of proinflammatory cytokines, such as IL-1β (28,29). Several studies have demonstrated the significance of miR-146a as a regulator of inflammation and NF-κB activity in vitro and in vivo (30,31). Moreover, miR-146a-5p has been confirmed to inhibit STAT3 activation (32). The present findings demonstrated that M2-exos and mimic-miR-146a-5p could reduce the protein levels of p-NF-κB p65 and p-STAT3, as determined by western blotting. Furthermore, double-immunofluorescence staining identified that pacing HL-1 myocytes treated with M2-exos and mimic-miR-146a-5p had reduced NF-κB expression. The current findings revealed that M2-exos and miR-146a-5p could reduce inflammation in pacing HL-1 myocytes by inhibiting NF-κB and STAT3.
NF-κB is a transcription factor that controls a wide range of biological reactions (33) and has been demonstrated to contribute to myocardial oxidative damage, inflammation and atrial remodeling in AF after myocardial infarction (34). In postoperative AF patients, STAT3 levels are increased in the atria, which promotes the fibrotic substrate and increases susceptibility to AF (35). Previous research has demonstrated that activated NF-κB enters the nucleus and controls the transcription of KCa3.1 genes (36,37) and suppressing the NF-κB pathway help to alleviate the inflammatory effect of KCa3.1 (38). In the present study, it was found that pacing HL-1 cells had increased protein levels of p-NF-κB p65, KCa3.1 and p-STAT3 via western blotting. The findings revealed that p-NF-κB p65 and p-STAT3 were crucial in the inflammatory response of tachypacing HL-1 myocytes. A recent study revealed that the KCa3.1 channel regulated the production of proinflammatory proteins via STAT3 (39) and that inhibition of KCa3.1 reduced STAT3 phosphorylation in a dose-dependent manner (38). In addition, early research revealed the interaction between STAT3 and NF-κB and explained how activating NF-κB signaling caused the production of IL-6, which then activated STAT3 (40). In the present study, inhibiting the NF-κB pathway with PDTC reduced the protein levels and current density of KCa3.1 and STAT3 and inhibited the shorter APD caused by rapid pacing. The current results confirmed that suppressing NF-κB might prevent STAT3 from being activated during tachypacing, leading to a decrease in KCa3.1 expression. Thus, M2-exos and mimic-miR-146a-5p could inhibit the KCa3.1 channel by targeting the NF-κB/STAT3 pathway.
In addition, both NF-κB and STAT3 are transcription factors that regulate IL-1β production by binding to the IL-1β promoter (41). In the present study, it was identified that blocking NF-κB in pacing HL-1 cells reduced the protein level of IL-1β. Pacing HL-1 myocytes that were treated with mimic-miR-146a-5p and M2-exos also reduced protein level of IL-1β. M2-exos and mimic-miR-146a-5p inhibited the NF-κB/STAT3 pathway, resulting in reduced IL-1β expression in pacing HL-1 myocytes.
In conclusion, rapidly paced HL-1 myocytes had increased KCa3.1 protein levels and current density, as well as the higher protein levels of NF-κB, STAT3, and IL-1β, and a shorter APD. Pacing HL-1 myocytes treated with M2-exo and miR-146a-5p mimic decreased the protein levels of NF-κB, STAT3 and IL-1β, and reduced KCa3.1 protein levels and current density, further reversing the shorter APD caused by rapid pacing. Inhibiting the NF-κB pathway produced a similar effect. The current findings demonstrated that M2-exos and their cargo, which encapsulated miR-146a-5p, may decrease KCa3.1 expression, and IL-1β secretion via the NF-κB/STAT3 signaling pathway, further ameliorating the shorter APD caused by pacing HL-1 cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81970277 and 82170312).
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
HC provided substantial contributions in the conceptualization, methodology, validation, investigation, writing of the original draft, and reviewing and editing. QY and MY were involved in designing this study, revising the manuscript critically for important intellectual content, and took charge of all aspects of the work to ensure that questions related to accuracy were resolved. HL and YF were assigned to the conceptualization and methodology. YY and DL confirm the authenticity of all the raw data, and were responsible for validation and investigation. ZC and ZP carried out the investigation and formal analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved (approval no. WDRM 20191211) by The Animal Research Subcommittee of the Renmin Hospital of Wuhan University Institutional Review Board, and complied with the standards established by the National Institutes of Health for the care and use of laboratory animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Hua T, Yang M, Song H, Kong E, Deng M, Li Y, Li J, Liu Z, Fu H, Wang Y and Yuan H: Huc-MSCs-derived exosomes attenuate inflammatory pain by regulating microglia pyroptosis and autophagy via the miR-146a-5p/TRAF6 axis. J Nanobiotechnology. 20:3242022. View Article : Google Scholar : PubMed/NCBI | |
|
Yuan Y, Mei Z, Qu Z, Li G, Yu S, Liu Y, Liu K, Shen Z, Pu J, Wang Y, et al: Exosomes secreted from cardiomyocytes suppress the sensitivity of tumor ferroptosis in ischemic heart failure. Signal Transduct Target Ther. 8:1212023. View Article : Google Scholar : PubMed/NCBI | |
|
Lin YN, Mesquita T, Sanchez L, Chen YH, Liu W, Li C, Rogers R, Wang Y, Li X, Wu D, et al: Extracellular vesicles from immortalized cardiosphere-derived cells attenuate arrhythmogenic cardiomyopathy in desmoglein-2 mutant mice. Eur Heart J. 42:3558–3571. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Sagris M, Vardas EP, Theofilis P, Antonopoulos AS, Oikonomou E and Tousoulis D: Atrial fibrillation: Pathogenesis, predisposing factors, and genetics. Int J Mol Sci. 23:62021. View Article : Google Scholar : PubMed/NCBI | |
|
Pegtel DM and Gould SJ: Exosomes. Annu Rev Biochem. 88:487–514. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Chen P, Wang L, Fan X, Ning X, Yu B, Ou C and Chen M: Targeted delivery of extracellular vesicles in heart injury. Theranostics. 11:2263–2277. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Shaihov-Teper O, Ram E, Ballan N, Brzezinski RY, Naftali-Shani N, Masoud R, Ziv T, Lewis N, Schary Y, Levin-Kotler LP, et al: Extracellular vesicles from epicardial fat facilitate atrial fibrillation. Circulation. 143:2475–2493. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang Z, Xu Y, Cao C, Wang B, Guo J, Qin Z, Lu Y, Zhang J, Zhang L, Wang W, et al: Exosomes as a messager to regulate the crosstalk between macrophages and cardiomyocytes under hypoxia conditions. J Cell Mol Med. 26:1486–1500. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Yao Y, He S, Wang Y, Cao Z, Liu D, Fu Y, Chen H, Wang X and Zhao Q: Blockade of exosome release suppresses atrial fibrillation by alleviating atrial fibrosis in canines with prolonged atrial pacing. Front Cardiovasc Med. 8:6991752021. View Article : Google Scholar : PubMed/NCBI | |
|
He S, Wang Y, Yao Y, Cao Z, Yin J, Zi L, Chen H, Fu Y, Wang X and Zhao Q: Inhibition of KCa3.1 channels suppresses atrial fibrillation via the attenuation of macrophage pro-inflammatory polarization in a canine model with prolonged rapid atrial pacing. Front Cardiovasc Med. 8:6566312021. View Article : Google Scholar : PubMed/NCBI | |
|
Sun Z, Ying X, Zhao W, He Y, Wang Z, Zheng L, Chen W and Xu H: M2c macrophages prevent atrial fibrillation in association with the inhibition of KCNQ1 in human embryonic stem cell-derived atrial-like cardiomyocytes. Hellenic J Cardiol. 62:457–459. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, et al: Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 7:15357502018. View Article : Google Scholar : PubMed/NCBI | |
|
Liu D, Yang M, Yao Y, He S, Wang Y, Cao Z, Chen H, Fu Y, Liu H and Zhao Q: Cardiac fibroblasts promote ferroptosis in atrial fibrillation by secreting Exo-miR-23a-3p Targeting SLC7A11. Oxid Med Cell Longev. 2022:39614952022.PubMed/NCBI | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Odell ID and Cook D: Immunofluorescence techniques. J Invest Dermatol. 133:e42013. View Article : Google Scholar : PubMed/NCBI | |
|
Al-Owais MM, Hettiarachchi NT, Dallas ML, Scragg JL, Lippiat JD, Holden AV, Steele DS and Peers C: Inhibition of the voltage-gated potassium channel Kv1.5 by hydrogen sulfide attenuates remodeling through S-nitrosylation-mediated signaling. Commun Biol. 6:6512023. View Article : Google Scholar : PubMed/NCBI | |
|
Yang Z, Shen W, Rottman JN, Wikswo JP and Murray KT: Rapid stimulation causes electrical remodeling in cultured atrial myocytes. J Mol Cell Cardiol. 38:299–308. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Brundel BJ, Kampinga HH and Henning RH: Calpain inhibition prevents pacing-induced cellular remodeling in a HL-1 myocyte model for atrial fibrillation. Cardiovasc Res. 62:521–528. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Weisbrod D, Khun SH, Bueno H, Peretz A and Attali B: Mechanisms underlying the cardiac pacemaker: The role of SK4 calcium-activated potassium channels. Acta Pharmacologica Sinica. 37:82–97. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Liebau S, Tischendorf M, Ansorge D, Linta L, Stockmann M, Weidgang C, Iacovino M, Boeckers T, von Wichert G, Kyba M and Kleger A: An inducible expression system of the calcium-activated potassium channel 4 to study the differential impact on embryonic stem cells. Stem Cells Int. 2011:4568152011. View Article : Google Scholar : PubMed/NCBI | |
|
Weisbrod D, Peretz A, Ziskind A, Menaker N, Oz S, Barad L, Eliyahu S, Itskovitz-Eldor J, Dascal N, Khananshvili D, et al: SK4 Ca2+ activated K+ channel is a critical player in cardiac pacemaker derived from human embryonic stem cells. Proc Natl Acad Sci USA. 110:E1685–E1694. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Haron-Khun S, Weisbrod D, Bueno H, Yadin D, Behar J, Peretz A, Binah O, Hochhauser E, Eldar M, Yaniv Y, et al: SK4 K channels are therapeutic targets for the treatment of cardiac arrhythmias. EMBO Mol Med. 9:415–429. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Yang M, Wang Y, Zhao H, Yin J, Zi L, Wang X, Tang Y, Huang C and Zhao Q: Role of intermediate-conductance calcium-activated potassium channels in atrial fibrillation in canines with rapid atrial pacing. J Interv Card Electrophysiol. 60:247–253. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Davidson SM, Boulanger CM, Aikawa E, Badimon L, Barile L, Binder CJ, Brisson A, Buzas E, Emanueli C, Jansen F, et al: Methods for the identification and characterization of extracellular vesicles in cardiovascular studies: From exosomes to microvesicles. Cardiovasc Res. 119:45–63. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Hirai K, Ousaka D, Fukushima Y, Kondo M, Eitoku T, Shigemitsu Y, Hara M, Baba K, Iwasaki T, Kasahara S, et al: Cardiosphere-derived exosomal microRNAs for myocardial repair in pediatric dilated cardiomyopathy. Sci Transl Med. 12:eabb33362020. View Article : Google Scholar : PubMed/NCBI | |
|
Dong M, Chen D, Zhu Y, Yang S, Kumar S, Zhang R, Zhou Y, Yang Z, Zheng N, Zhu T, et al: Impaired regulation of MMP2/16-MLCK3 by miR-146a-5p increased susceptibility to myocardial ischaemic injury in aging mice. Cardiovasc Res. 119:786–801. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Rivera-Caravaca JM, Teruel-Montoya R, Roldán V, Cifuentes-Riquelme R, Crespo-Matas JA, de Los Reyes-García AM, Águila S, Fernández-Pérez MP, Reguilón-Gallego L, Zapata-Martínez L, et al: Pilot study on the role of circulating miRNAs for the improvement of the predictive ability of the 2MACE score in patients with atrial fibrillation. J Clin Med. 9:36452020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang QB, Qing YF, Yin CC, Zhou L, Liu XS, Mi QS and Zhou JG: Mice with miR-146a deficiency develop severe gouty arthritis via dysregulation of TRAF 6, IRAK 1 and NALP3 inflammasome. Arthritis Res Ther. 20:452018. View Article : Google Scholar : PubMed/NCBI | |
|
Pan J, Du M, Cao Z, Zhang C, Hao Y, Zhu J and He H: miR-146a-5p attenuates IL-1β-induced IL-6 and IL-1β expression in a cementoblast-derived cell line. Oral Dis. 26:1308–1317. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Su YL, Wang X, Mann M, Adamus TP, Wang D, Moreira DF, Zhang Z, Ouyang C, He X, Zhang B, et al: Myeloid cell-targeted miR-146a mimic inhibits NF-κB-driven inflammation and leukemia progression in vivo. Blood. 135:167–180. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Meng Q, Liang C, Hua J, Zhang B, Liu J, Zhang Y, Wei M, Yu X, Xu J and Shi S: A miR-146a-5p/TRAF6/NF-kB p65 axis regulates pancreatic cancer chemoresistance: Functional validation and clinical significance. Theranostics. 10:3967–3979. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Sun W, Ma J, Zhao H, Xiao C, Zhong H, Ling H, Xie Z, Tian Q, Chen H, Zhang T, et al: Resolvin D1 suppresses pannus formation via decreasing connective tissue growth factor caused by upregulation of miRNA-146a-5p in rheumatoid arthritis. Arthritis Res Ther. 22:612020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang L, Wei X, Wang Z, Liu P, Hou Y, Xu Y, Su H, Koci MD, Yin H and Zhang C: NF-κB activation enhances STING signaling by altering microtubule-mediated STING trafficking. Cell Rep. 42:1121852023. View Article : Google Scholar : PubMed/NCBI | |
|
Liu X, Zhang W, Luo J, Shi W, Zhang X, Li Z, Qin X, Liu B and Wei Y: TRIM21 deficiency protects against atrial inflammation and remodeling post myocardial infarction by attenuating oxidative stress. Redox Biol. 62:1026792023. View Article : Google Scholar : PubMed/NCBI | |
|
Liu Y, Wu F, Wu Y, Elliott M, Zhou W, Deng Y, Ren D and Zhao H: Mechanism of IL-6-related spontaneous atrial fibrillation after coronary artery grafting surgery: IL-6 knockout mouse study and human observation. Transl Res. 233:16–31. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Ye T, Zhang C, Wu G, Wan W, Liang J, Liu X, Liu D and Yang B: Pinocembrin attenuates autonomic dysfunction and atrial fibrillation susceptibility via inhibition of the NF-κB/TNF-α pathway in a rat model of myocardial infarction. Int Immunopharmacol. 77:1059262019. View Article : Google Scholar : PubMed/NCBI | |
|
Xu H, Lai W, Zhang Y, Liu L, Luo X, Zeng Y, Wu H, Lan Q and Chu Z: Tumor-associated macrophage-derived IL-6 and IL-8 enhance invasive activity of LoVo cells induced by PRL-3 in a KCNN4 channel-dependent manner. BMC Cancer. 14:3302014. View Article : Google Scholar : PubMed/NCBI | |
|
Zheng F, Tao Y, Liu J, Geng Z, Wang Y, Wang Y, Fu S, Wang W, Xie C, Zhang Y and Gong F: KCa3.1 inhibition of macrophages suppresses inflammatory response leading to endothelial damage in a cell model of kawasaki disease. J Inflamm Res. 14:719–735. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Jiang XX, Bian W, Zhu YR, Wang Z, Ye P, Gu Y, Zhang H, Zuo G, Li X, Zhu L, et al: Targeting the KCa3.1 channel suppresses diabetes-associated atherosclerosis via the STAT3/CD36 axis. Diabetes Res Clin Pract. 185:1097762022. View Article : Google Scholar : PubMed/NCBI | |
|
McFarland BC, Hong SW, Rajbhandari R, Twitty GB Jr, Gray GK, Yu H, Benveniste EN and Nozell SE: NF-κB-induced IL-6 ensures STAT3 activation and tumor aggressiveness in glioblastoma. PLoS One. 8:e787282013. View Article : Google Scholar : PubMed/NCBI | |
|
Cogswell JP, Godlevski MM, Wisely GB, Clay WC, Leesnitzer LM, Ways JP and Gray JG: NF-kappa B regulates IL-1 beta transcription through a consensus NF-kappa B binding site and a nonconsensus CRE-like site. J Immunol. 153:712–723. 1994. View Article : Google Scholar : PubMed/NCBI |