Advancements in colorectal cancer research: Unveiling the cellular and molecular mechanisms of neddylation (Review)
- Authors:
- Published online on: February 21, 2024 https://doi.org/10.3892/ijo.2024.5627
- Article Number: 39
-
Copyright: © Wang et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
1. Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed type of cancer globally and ranks second in the cause of cancer-related mortality (1). It has a momentous effect on the lives and health of individuals. The 5-year survival rates are related to the stage at which the disease is detected; patients with stage I disease have a 5-year survival range >90%, whereas for patients with stage IV disease, the survival rate slightly exceeds 10% in clinical observations. Screening has been shown to reduce CRC morbidity and mortality (2). However, for patients in which the disease is not detected in time, follow-up treatment remains the sole method which allows them to continue to survive.
The majority of surgical treatments for patients with stage I and II CRC are based on surgical resection and the dissection of the surrounding lymph nodes. Apart from a small number of patients who develop post-operative intestinal knotting and wound dehiscence, the majority have a good prognosis and 5-year survival rates. At present, the majority of patients who are eligible for surgical treatment, based on health status and the stage of the disease (3,4), receive open surgery (5). Simultaneously, minimally invasive surgeries, such as laparoscopic surgeries (6,7) and robotic surgeries (8) have gradually gained a place in surgical treatment options due to their characteristics of diminished hemorrhagic tendencies, expedited post-operative recuperation, and lighter immune and inflammatory reactions than open surgeries.
Patients with stage III CRC frequently undergo chemotherapy to mitigate the risk of recurrence. In tandem, individuals with stage II or III CRC for whom surgical interventions prove intolerable may resort to a combined approach, where chemotherapy is complemented by radiation therapy. For patients with stage IV disease, the main treatment option is chemotherapy. Furthermore, adjuvant chemotherapy, peri-operative chemotherapy (9), and neoadjuvant chemoradiotherapy (10,11) not only reduce the disease stage, but also curtail the likelihood of recurrence and elevate the overall survival prospects for these patients. However, hepatotoxicity caused by chemotherapeutic drugs (12), particularly in frail or susceptible elderly patients (13), limits their application.
An increasing number of targeted therapies are also being applied for the treatment of CRC, which significantly enhance the prognosis of patients with malignant tumors compared to other treatments, particularly in individuals diagnosed with metastatic CRC (3,13). Currently, the most frequently used targeted therapies include angiogenesis inhibitors, such as bevacizumab and epidermal growth factor (EGF) inhibitors, such as panitumumab and cetuximab, alongside BRAF inhibitory therapy and HER2 inhibitory therapy (14). Neural precursor cell-expressed developmentally downregulated 8 (NEDD8) has attracted widespread attention for its function in CRC. CRC tissues exhibit an elevated expression of NEDD8, and the participation of NEDD8 in neddylation plays a crucial role in the migration, proliferation and survival of cancer cells. The present review summarizes and discusses the influence and potential of targeted NEDD8 therapy in CRC.
2. Overview of neddylation
NEDD8 was first discovered in mouse neural precursor cells (15) encoding a protein characterized by an arrangement of 81 amino acid residues. The molecular structure of NEDD8 is similar to that of ubiquitin, with up to 60% shared identity (16) and 80% homology (17). Furthermore, it is worth highlighting that the C-terminus of the protein encoded by NEDD8 comprises an uninterrupted quartet of Gly residues, with remote residues such as Gly-75 and Gly-76 bearing the hallmark of evolutionary conservation. The study by Kamitani et al (17) confirmed that only Gly-76 is related to the formation of NEDD8 conjugates, and this characteristic exhibits a resemblance to ubiquitin. Moreover, both ubiquitin and NEDD8 share conserved amino acid residues, such as Lys-48. The process of combining NEDD8 with other proteins is similar to ubiquitination, although the mechanism of binding markedly diverges from that of ubiquitin. Furthermore, unlike ubiquitin, NEDD8 assumes a critical role in orchestrating the modulation of linked signal transduction, transcription factors, tumorigenesis and other biological processes.
The neddylation process
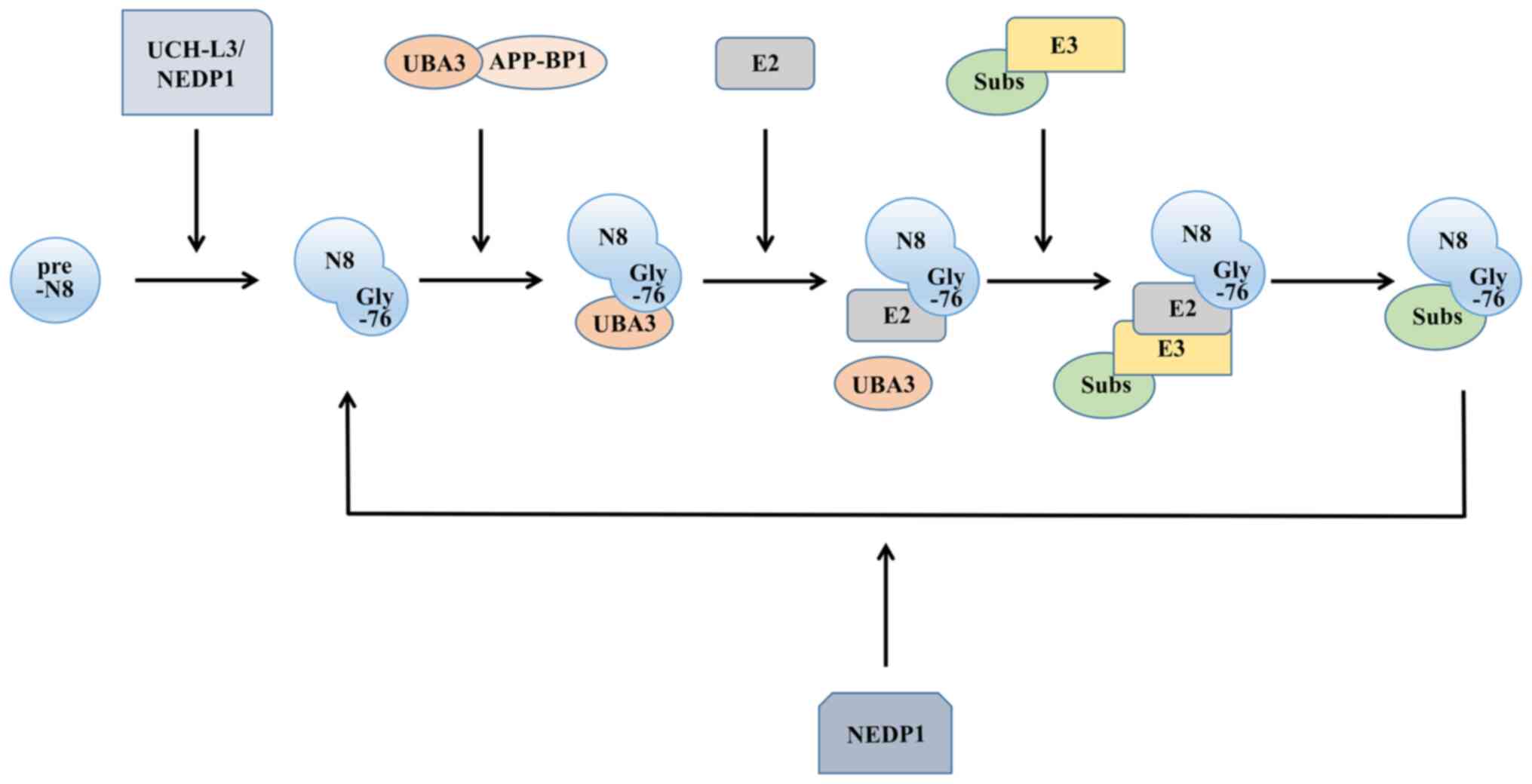
As a post-translational modification mechanism for proteins, neddylation shares a number of similarities with ubiquitination. There are four main enzymes involved in the ubiquitin-like process: A precursor processing enzyme, NEDD activating enzyme (E1), NEDD conjugating enzyme (E2) and NEDD ligase (E3). The NEDD8 molecule is synthesized in a precursor form. To expose the Gly-76 residue, the C-terminus of the protein undergoes cleavage (17), where the precursor processing enzymes activate, including ubiquitin carboxyl-terminal hydrolase isozyme L3 (18), a common precursor processing enzyme for NEDD8 and ubiquitin, and the NEDD8-specific precursor processing enzyme NEDD8 protease 1 (19).
After processing the precursor, NEDD8 still needs to be activated to function, similar to ubiquitin, and this process is completed by E1. Functioning as a heterodimer, it comprises both ubiquitin-like modifier activating enzyme 3 and amyloid beta precursor protein binding protein 1. E1 then transfers NEDD8 to the thiol lipid coupling intermediate through a transesterification reaction (20). This forms the E2-NEDD8 conjugates. The E2s discovered thus far mainly include ubiquitin-conjugating enzyme (UBE)2M [also known as ubiquitin-conjugating enzyme C12 (UBC12)] (21) and UBE2F (22). Finally, E3, with the function of determining the specificity of the substrate, is required to localize the NEDD8 molecule to the target molecule. Compared with ubiquitination, neddylation has fewer types of E3s (Fig. 1).
NEDD8 exhibits specificity in binding to substrate proteins. E3 ligases, in conjunction with their respective E2 binding enzymes, typically collaborate in recruiting specific protein substrates to the substrate receptor domain of E3. NEDD8 then undergoes transfer from the independent active site of E3, ultimately facilitating the interaction between NEDD8 and the specific substrate proteins. For instance, ring box protein 1 demonstrates the capacity to use either UBE2M or UBE2F, affecting the binding of NEDD8 to the specific lysine residue 720 within cullin 1 (23). E2 also affects the binding site of E3, which prevents inappropriate modifications from occurring (21).
Neddylation substrate
There are numerous substrates for neddylation, among which the most characterized and well-documented are the cullin family proteins (24). In addition, other typical substrates are p53 (25), the Von Hippel-Lindau tumor suppressor protein (pVHL) (26), proliferating cell nuclear antigen (PCNA) (27), hypoxia-inducible factor (HIF) (28), and murine double minute 2 (MDM2) (25).
Cullin family proteins
Molecular scaffolds of cullin-ring ligase (CRL) E3 ubiquitin ligases involve cullin family proteins (29), which serve as adaptable frameworks, connecting the active site of E2 enzymes with the substrate binding site and facilitating the progression of the ubiquitination process by bridging the spatial gap (30). There are seven types of cullins in mammals: Cullin1, cullin2, cullin3, cullin4A, cullin4B, cullin5 and cullin7 (31), of which cullin4A was the first to be discovered (32). A crucial characteristic of CRL E3 is its dependence on the covalent modification of the cullin protein by the NEDD8 molecule for its activation, resulting in a relatively active conformation of the ring-type E3 ligase (33). Additionally, CRL E3 ubiquitin ligases play crucial roles in coordinating the conjugation of ubiquitin with substrate proteins. They are able to bind to the target proteins and recruit E2 enzymes that are responsible for the transfer of ubiquitin molecules (31). The neddylation of cullin proteins can then instigate a profound transformation in their conformation, promoting neddylation of the CRL substrate.
MDM2/p53
p53 is a tumor suppressor gene with transcription factor activity related to DNA damage repair. In the case that its function is inhibited, the capacity to efficiently mend DNA damage is compromised, leading to its unabated accumulation. This, in turn, paves the way for genome instability, ultimately fostering the emergence of tumorigenesis. MDM2, a ring-type E3 ligase, is mainly amplified in cancer. Notably, it holds the power to catalyze the neddylation of p53, effectively curtailing the transcriptional prowess of this vital transcription factor. At the same time, MDM2 can also undergo neddylation itself, thereby increasing its stability and catalyzing p53 to undergo neddylation (25). Consequently, the functionality of p53 is suppressed, leading to heightened genomic instability within the malignant cells, thereby facilitating the progression of tumor growth.
pVHL/HIF
HIF-1α, which facilitates cancer development by enhancing angiogenesis (34), is an oxygen-regulated transcriptional activator (35). pVHL is one of the components of the E3 ubiquitin ligase that ubiquitinates and degrades hydroxylated HIF-1α. The functions of ubiquitination and degradation of hydroxylated HIF-1α remain unaltered following neddylation, which concurrently affects tumor development by affecting the assembly of the fibrin matrix (26). Under hypoxic conditions, the HIF-1α concentration is maintained by neddylation so that there is oxidative stress in cells, thereby promoting the development of tumor cells (28).
PCNA
PCNA is involved in DNA synthesis and cell proliferation. The molecular interplay between PCNA and NEDD8 possesses the capacity to counteract the process of ubiquitination. However, PCNA that undergoes neddylation blocks the formation of polη lesions, thereby inhibiting the recruitment of polη and affecting DNA damage repair, which may cause genome instability and lead to tumor occurrence (27).
Role of neddylation in tumorigenesis
Neddylation is related to tumor proliferation
Irregularities within the normal cellular replication process cause the body to lose normal control of its cell cycle and to prompt anomalous cell proliferation, -a decisive sign of tumorigenesis (36). The deficiency of enzymes involved in the neddylation process can result in the inhibition of tumor proliferation. For example, MLN4924, which selectively inhibits the E1 regulatory subunit [NEDD activating enzyme (NAE)], can interfere with neddylation in a number of types of cancer (37-39), particularly CRC (40), leading to cell cycle arrest or the demise of the cell. In addition, SMAD ubiquitination regulatory factor (Smurf)1, functioning as an E3 ligase, promotes its own neddylation, a process which can subsequently enhance CRC proliferation (41).
Neddylation is related to tumor metastasis
Metastasis encompasses the dissemination of cancer cells from the initial location to neighboring tissues or other organs in the body and their proliferation in the post-metastasis site, which is the crucial cause of tumor occurrence and mortality (36). MLN4924, an inhibitor of neddylation, impedes the metastatic journey of neoplastic cells by thwarting their intravascular persistence and the intricate extravasation process (42). NEDD8, via the s-phase kinase associated protein 2 (Skp2)/Slug pathway, exerts a regulatory effect on the downregulation of E-cadherin (43), consequently triggering the activation of the epithelial-to-mesenchymal transition (EMT) (44).
Neddylation is associated with tumor resistance to cell death
Resisting cell death is one of the abilities of tumors (36), contributing to the enhanced survival of cancer cells under adverse conditions. p73 and its isoforms play a pivotal role in promoting apoptosis in the cell cycle. NEDD8 can attenuate the transactivation of Tap73 through the MDM2/Tap73 pathway (45), as well as reduce the activation of p73 through the E2F transcription factor 1 (E2F1)/p73 route, allowing E2F1 to function in promoting the activation of cell cycle progression genes (46). Double-strand breaks (DSBs) in DNA present the most crucial issue for cells among all DNA damage (47). NEDD8 can facilitate the repair of DSBs (48), with the key player in the repair process being the COP9 signalosome (CSN). DSBs sites recruit CSN through neddylation and mainly interact with cullin4A, which is necessary for DSB repair (49). This DSB repair allows cells to escape death caused by genetic damage. Additionally, NEDD8 is intricately linked to resistance to tumor treatment (50). Resistance to treatment is also a form of tumor resistance to cell death and is closely related to drug resistance and recurrence (51). It has been shown that targeting neddylation enhances the sensitivity of CRC to the topoisomerase I inhibitor in chemotherapy (52). MLN4924 emerges as a novel radiosensitizer, augmenting the sensitivity of CRC to radiotherapy (53). In addition, the interaction between circAFF2 and cullin-associated and neddylation-dissociated protein 1 (CAND1) facilitates the binding of CAND1 to cullin5, suppressing cullin5 neddylation and consequently enhancing radiosensitivity in CRC (54).
Neddylation is related to the tumor microenvironment
Tumor-induced interactions dominate the tumor microenvironment (55). In myeloid-derived suppressor cells, NEDD8 can enhance the enrichment of tumor-infiltrating immune cells, contributing to the creation of an immunosuppressive environment (55,56). NEDD8, by virtue of its role in orchestrating the degradation of IκB via the cullin1-mediated pathway, activates NF-κB, subsequently elevating the release of particular pro-inflammatory cytokines induced by lipopolysaccharides, such as IL-6 and TNF-α (57). This is closely related to the role of tumor-associated macrophages (TAMs) (58). Likewise, the suppression of neddylation significantly inhibits the infiltration of TAMs (59). The anticancer immune response is significantly influenced by the immune response triggered through the activation of T-cells (60). Silencing the expression of UBC12 leads to a marked reduction in neddylation, targeting the adapter protein, src homology collagen, within CD4+ T-cells. This causes a discernible impairment in cytokine production simultaneous to a diminished activation of the extracellular signal-regulated kinase (ERK) (61). At the same time, the targeted inhibition of neddylation can not only reduce NF-κB activity (62), but can also regulate the polarization of T-cells in vitro (63). Tumor angiogenesis is a necessary condition for tumor progression (64) and is mainly associated with vascular endothelial growth factor (VEGF) (65). It has been found that the UBC12 concentration increases in accordance with VEGF concentration (66). Furthermore, CRL can be inhibited by MLN4924, culminating in the accumulation of early rat sarcoma homolog gene family A (RhoA) and detrimentally impacting the angiogenic prowess of vascular endothelial cells (67) (Table I).
3. The function of neddylation in colorectal cancer
NEDD8 is inseparable from animal development and plays a crucial regulatory role in cell metastasis and proliferation. In addition, it is related to cell survival and genetic changes. Previous research has confirmed that UBC12, highly expressed in individuals with CRC, is involved in the activation pathway of NEDD8. This upregulation in the expression of NEDD8 is related to tumor cell proliferation and cloning ability (41).
CRC remains a significant contributor to cancer-related mortality. Although patients can achieve certain control of the disease through surgical resection and chemotherapy, the majority of patients eventually succumb to the disease due to tumor invasion and metastasis (68). The role of neddylation in CRC primarily lies in fostering the proliferation and migration of tumor cells, while concurrently protecting these cells from apoptotic processes. It is evident that NEDD8 intricately intertwines with the course of numerous cancers, including CRC, and delving into the association between NEDD8 and CRC illustrates paramount significance.
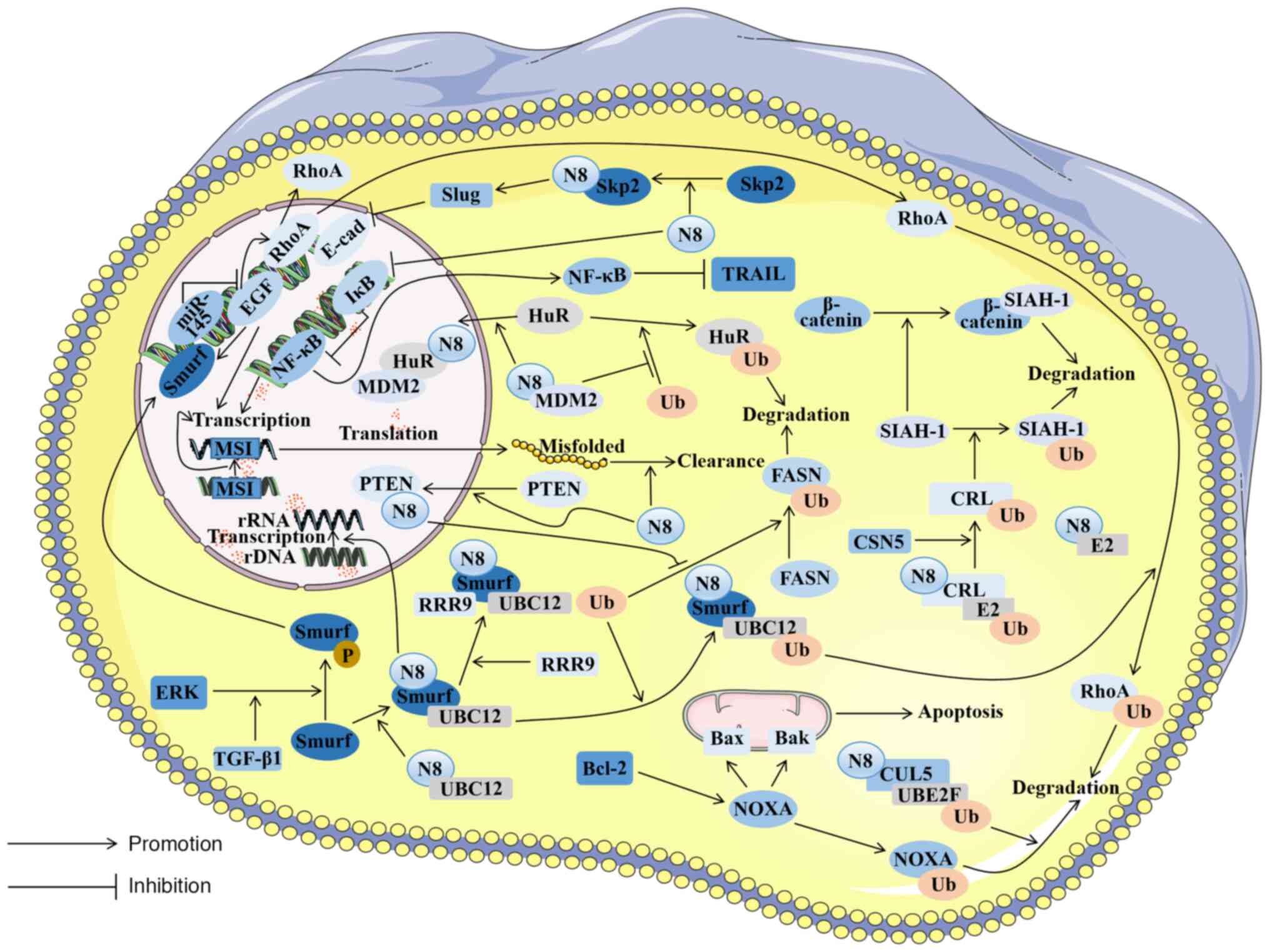
NEDD8 promotes tumor cell proliferation and metastasis through Smurf/E2 and Smurf/ribosomal RNA processing 9 (RRP9)
NEDD8 functions by covalently binding to substrates in an ubiquitination-like manner (69). Its E3 not only has a ring scaffold type, but also involves a HECT ligase. Smurf1 emerges as a C2-WW-HECT ligase. Moreover, Smurf1 serves as a ubiquitin ligase E3, whose expression is substantially heightened within CRC tissues (41). Furthermore, in CRC tissues, increased levels of Smurf1, NEDD8, NAE1 and UBC12 expression have been linked to the advancement of cancer and to unfavorable clinical outcomes (41).
Smurf1 can be activated through neddylation, promoting its association with E2. Additionally, Smurf1 possesses the capacity to serve as an E3, thereby catalyzing its own neddylation process (41). The indispensable roles of NEDD8 and Smurf1 are evident in facilitating the ubiquitination of RhoA, a protein mainly related to the migration of tumor cells (70). In breast cancer (71), elevated Smurf1 expression has been substantiated as a consequence of ERK-mediated phosphorylation, which results from the activation of transforming growth factor β1 (TGF-β1) (72). It is worth noting that the overexpression and activation of ERK have also been found in CRC (73). Hence, it is conceivable that a similar mechanism is enacted in CRC. However, some researchers have illustrated that the expression of RhoA is increased in CRC due to EGF (74).
EGF plays a critical role in tumor cell metastasis (75). The downregulation of microRNA (miR)-145, leading to the upregulation of EGF, has been detected in CRC (76). At the same time, it has been confirmed that EGF can promote Smurf1 expression (77), thereby facilitating the neddylation of Smurf1 and prompting TGF-β1-induced ERK to phosphorylate Smurf1, ultimately resulting in the ubiquitination and degradation of RhoA. This appears contradictory to the previously mentioned elevation of RhoA induced by EGF (74). However, during cell migration, RhoA exhibits disparate functionalities in the anterior and posterior regions of tumor cells. The former is mainly inhibited and the latter is mainly activated (78). In CRC, the EFG-induced accumulation of RhoA may be intricately associated with the posterior section of migrating cells. The pathway responsible for diminishing RhoA ubiquitination, mediated by NEDD8, could potentially be linked to the migration of the front of the cell. In addition, the neddylation of Smurf1 can control rRNA preprocessing to promote tumor cell proliferation. As a result, cell proliferation and migration experience severe inhibition in cells, in turn knocking out the Smurf1 gene (79). In addition to the neddylation of Smurf, the mapping of the dynamic signaling network of TGF-β using high-throughput techniques has revealed that RRP9 can interact with Smurf1 (80) as a Smurf1 substrate in neddylation. In human CRC tissue samples and matched adjacent normal tissue samples, significantly upregulated expression levels of RRP9 and Smurf1 have been found in CRC tissues (79).
Substantiated investigations have elucidated the interconnection of RRP9 and NEDD8 through Smurf1 in CRC (79). RRP9, the integral element within the U3 snoRNP complex, augments pre-rRNA processing via its ubiquitination (79,81), an indispensable step in ribosome biogenesis. This augmentation in ribosome biogenesis is frequently associated with the increased proliferation observed in cancer as a high production of ribosomes is necessary to maintain high levels of cell growth and division (82,83). Therefore, Smurf1 exerts its E3 function and not only catalyzes RRP9 neddylation, but also stimulates the proliferation and migration of CRC cells by increasing pre-rRNA processing and upregulating ribosome biogenesis (79). Therefore, Smurf1 exhibits an elevated expression level and is activated through the process of neddylation, promoting the development of colorectal tumors. In the realm of CRC treatment, studies have illuminated that the inhibition of Smurf1 in tumors manifests an amplified anti-tumor efficacy in response to gemcitabine, cisplatin, and the gemcitabine-cisplatin combination (84). This observation suggests that addressing chemotherapy-resistant CRC by therapeutically targeting NEDD8 to suppress Smurf1 activation holds substantial promise.
NEDD8/X-linked inhibitor of apoptosis protein (XIAP)/phosphatase and tensin homolog (PTEN)/fatty acid synthase (FASN) increases tumor cell proliferation by promoting fat synthesis
PTEN encodes a protein phosphatase with strong tumor inhibitory effects (85). It is distributed in both the nucleus and cytoplasm and has complementary mechanisms to inhibit tumor development (86). PTEN has been documented as a substrate susceptible to neddylation, capable of forming a conjugate with NEDD8 by means of XIAP. Moreover, it has been found that, in breast cancer tissue, PTEN neddylation is triggered by increases in glucose levels (87), which is reflected in the process of cancer development. Because the propagation of cancerous cells hinges on the acquisition of extracellular nutrients (88), including glucose (89), research has shown that individuals with hyperglycemia are more susceptible to the onset of CRC. Furthermore, an extreme stimulation of glucose uptake and glycolytic activity stands as a hallmark of CRC. This phenomenon is considered to be intricately linked to the surge in PTEN ubiquitination modification triggered by heightened sugar levels. XIAP is an E3 catalyzing PTEN neddylation that demonstrates an elevated expression level within CRC tissue and may mediate excessive PTEN neddylation.
It has been found that the effect of NEDD8 on PTEN is not derived from stabilizing its molecular structure, but from facilitating the translocation of PTEN into the nucleus in breast cancer cells. NEDD8 interacts with importin α, importin β and importin 5 (IPO5), leading to the nuclear translocation of any PTEN that has interacted with NEDD8 (87). Research on CRC cells has demonstrated that a high expression of IPO5 was associated with tumor development (90), suggesting that NEDD8 promotes PTEN nuclear translocation in CRC cells. NEDD8 stabilizes FASN through the NEDD8/XIAP/PTEN/FASN pathway, thereby reducing FASN ubiquitination and promoting fat synthesis (88). Elevated fat synthesis affected by FASN stabilization is often related to tumor cell proliferation (91). It has also been confirmed that Smurf1 can ubiquitinate and degrade PTEN in glioblastoma (92), which has not yet been proven in CRC. In the domain of breast cancer therapy, the targeted neddylation of PTEN has emerged as a highly promising therapeutic strategy (93). Building upon the analogous PTEN neddylation pathway, targeted interventions along this route hold significant developmental prospects in the context of CRC.
CSN5 promotes tumor cell proliferation by depleting NEDD8 to accumulate β-catenin
Abnormities in the Wnt/β-catenin signaling pathway are prevalent within the majority of CRC cells. The constitutive activation of this pathway promotes the propagation and survival of CRC cells and is a main cause of adenoma formation (94). Principal among these anomalies is the irregular expression of β-catenin, which serves as the primary instigator of the irregular signaling of the Wnt/β-catenin pathway in CRC cells. This perturbation exhibits an intricate association with CSN5. CSN5 mainly affects an alternative degradation pathway of β-catenin (95). In CRC cells, the intricate web of interactions exists among β-catenin, CSN5, and seven in absentia homolog 1 (SIAH-1), in which CSN5 plays a crucial role in cellular processes, such as apoptosis and cell cycle regulation. CRL activity can be regulated by deleting NEDD8 (96) to promote SIAH-1 degradation, causing β-catenin accumulation in CRC and promoting the growth and multiplication of malignant cells (97). Adenoma genesis frequently emerges as a pivotal risk factor in the onset of CRC (98). Consequently, the targeted development of pharmaceutical agents addressing this pathway holds the potential to contribute significantly to the prevention of CRC, thereby mitigating the incidence of this malignancy.
NEDD8 downregulates E-cadherin and activates EMT to promote tumor cell metastasis through the Skp2/Slug pathway
Research has illustrated that inhibiting the NEDD8 pathway has a therapeutic effect on clinically invasive CRC and that CRC cells exhibiting heightened sensitivity to NEDD8 inhibition often exhibit EMT transcriptional characteristics (40). EMT is an important prerequisite for CRC cells to metastasize (99), but the mechanisms of NEDD8 in inducing CRC cell metastasis have not been fully elucidated. In prostate cancer, which is also aggressive, it has been found that NEDD8 can down-regulate E-cadherin and activate EMT through the Skp2/Slug pathway to promote tumor cell metastasis (43). As a member of the substrate recognition F-box protein family, Skp2 is a component of CRL1 whose activity is regulated by NEDD8 (100). In other words, Skp2 depends on NEDD8 for its function. NEDD8 combines with Skp2 to perform neddylation, thereby activating the Skp2 downstream molecule Slug. Slug activation may suppress EMT onset by downregulating E-cadherin expression (101). Studies have found that both Skp2 (102) and Slug (103) exhibit a significant upregulation within tissues afflicted by CRC. Moreover, the downregulation of E-cadherin in these tissues also leads to the occurrence of EMT (104). It can be inferred that there is a mechanism in CRC tissues, whereby NEDD8 initiates the Skp2/Slug pathway, culminating in the downregulation of E-cadherin. This, in turn, prompts neoplastic cells to undergo the process of EMT, ultimately facilitating cellular metastasis.
NEDD8 mediates clearance of misfolded aggregates to protect tumor cells with microsatellite instability (MSI)
The process of DNA mismatch repair (MMR) is evolutionarily conserved and serves to rectify mismatches arising from DNA replication, evading proofreading mechanisms (105), in a key pathway in DNA repair. The loss of MMR function induces a hypermutational phenotype, and such cells are clinically identified through the genomic scarring of MSI. It has been confirmed that the prognosis of individuals with CRC is associated with the presence of MSI within the tumor (106). Patients with MSI develop intrinsic resistance to chemotherapy, making it difficult to induce cancerous cell apoptosis by chemotherapy drugs, limiting the number of effective treatments for patients (107). There are often a large number of genome instabilities and mutations in MSI, resulting in cellular proteome imbalances and aberrant protein accumulation in the cell. To compensate for these proteins, NEDD8-mediated pathways are indispensable for MSI tumors to clean misfolded aggregates, thereby maintaining tumor cell survival (108).
The NEDD8/UBE2F/cullin5 pathway strengthens NOXA ubiquitination and degradation to protect tumor cells
Defects in apoptosis are the basis of tumorigenesis and the main cause of chemotherapy failure (109). The role of the B-cell lymphoma 2 (Bcl-2) protein family in cellular apoptosis is of utmost importance, particularly in mitochondrial apoptosis (110). Within the cellular proteome, the Bcl-2 family can be segregated into two distinct categories: Pro-apoptotic and anti-apoptotic. The stoichiometry of these two categories determines whether the cell is apoptotic or not. When the subtle balance is altered, a signal is transmitted through the upstream molecule Bcl-2 homology domain only protein (BH3-only), thereby activating Bcl-2-associated X protein (Bax) and Bcl-2-associated K protein (Bak) on the mitochondrial surface, leading to mitochondrial impairment and cell death (111). NOXA is not only a pro-apoptotic protein, but is also a member of the BH3-only family. It plays a key role in Bcl-2-mediated mitochondrial apoptosis, mainly by selectively neutralizing anti-apoptotic Bcl-2 family proteins and altering the balance of pro- and anti-apoptotic signals to cause mitochondrial apoptosis (112). Research has indicated an elevated protein expression of NOXA in CRC; however, it has a short lifespan. The rapid degradation of NOXA could potentially serve as a tactic employed by CRC cells to evade the peril posed by elevated NOXA expression. Peroxiredoxin 1 exhibits a high expression level within CRC tissues, which enhances cullin5 neddylation through the UBE2F/cullin5 pathway. This neddylation can enhance the ubiquitination and degradation of NOXA mediated by cullin5, resulting in increased tumor cell survival, which is also reflected in the context of tumor cell resistance to the chemotherapy drug etoposide (113). Addressing this pathway through targeted interventions holds great promise in circumventing resistance to etoposide in CRC, thereby presenting a novel perspective for the chemotherapy of colorectal malignancies.
The NEDD8/MDM2/Hu antigen R (HuR) pathway stabilizes HuR to protect tumor cells
A central RNA-binding protein known as HuR plays a crucial role in stabilizing cell proliferation-related mRNA to regulate cell dedifferentiation, proliferation and survival (114,115). A previous study validated the significant association between the elevated expression of HuR and the survival of cancerous cells in tumors (116), while the decreased expression of HuR resulted in cell cycle arrest and promoted cellular apoptosis. A previous study detected the upregulation of HuR and MDM2 expression in colorectal tumor tissues (117). Notably, an intricate correlation has been established between HuR and MDM2. MDM2 has been previously confirmed to be an E3 that inhibits the transcriptional activity of p53 by catalyzing its neddylation (25). Moreover, MDM2 serves as an E3 in CRC tissues. The catalytic substrate of MDM2 is converted into HuR simultaneously, controlling the nuclear localization of HuR through the NEDD8/MDM2/HuR pathway and protecting it from degradation. To help tumor cells to survive, NEDD8 leads to the malignant transformation of tumor cells (118).
NEED8 protects tumor cells through the IκB/NF-κB pathway
Chemoresistance is often related to patient mortality and tumor metastasis in CRC; one form of chemoresistance is resistance to oxaliplatin through the death receptor ligand [Fas and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)] pathway (119). NF-κB plays a pivotal role in the regulation of death receptor signaling pathways (120). In chemoresistant diffuse large B-cell lymphoma, it has been found that neddylation resists tumor cell death by downregulating inhibitory factor IκB and upregulating NF-κB. MLN4924 can reverse such effects (121). The abnormal expression of NF-κB has also been detected in CRC cells (122). In an experiment involving the intraperitoneal injection of mutant IκB colon cells into mice suffering from metastatic CRC, it was discovered that this approach improved the survival rate of the mice, suggesting that NF-κB inhibition mediates the cell-killing effect caused by TRAIL (123). Similarly, NEDD8 is abnormally activated in CRC tissues. It can be inferred that NEDD8's presence in CRC inhibits IκB from activating the NF-κB pathway, thereby inhibiting TRAIL-mediated cell killing and allowing cancer cells to survive in the face of chemical drugs. In light of the deleterious ramifications stemming from cancer cells' resistance to platinum-based drugs, notably oxaliplatin, in the majority of CRCs (123), the targeted development of pharmaceutical agents along this pathway holds immense promise. This endeavor has the potential to furnish novel therapeutic strategies for treatment-resistant CRC (Fig. 2).
4. Exploration of neddylation as a potential molecularly targeted drug and of treatment for colorectal cancer
Neddylation is closely connected to the metastasis, proliferation and survival of CRC cells. Previous research has confirmed that inhibiting neddylation mediated by NEDD8 causes the apoptosis of CRC cells (124). Thus, targeting the neddylation pathway is expected to become a novel treatment method for CRC. Concurrently, such targeted therapy may also find application in combination with CRC treatment drugs to improve the responsiveness of CRC cells to therapeutic agents.
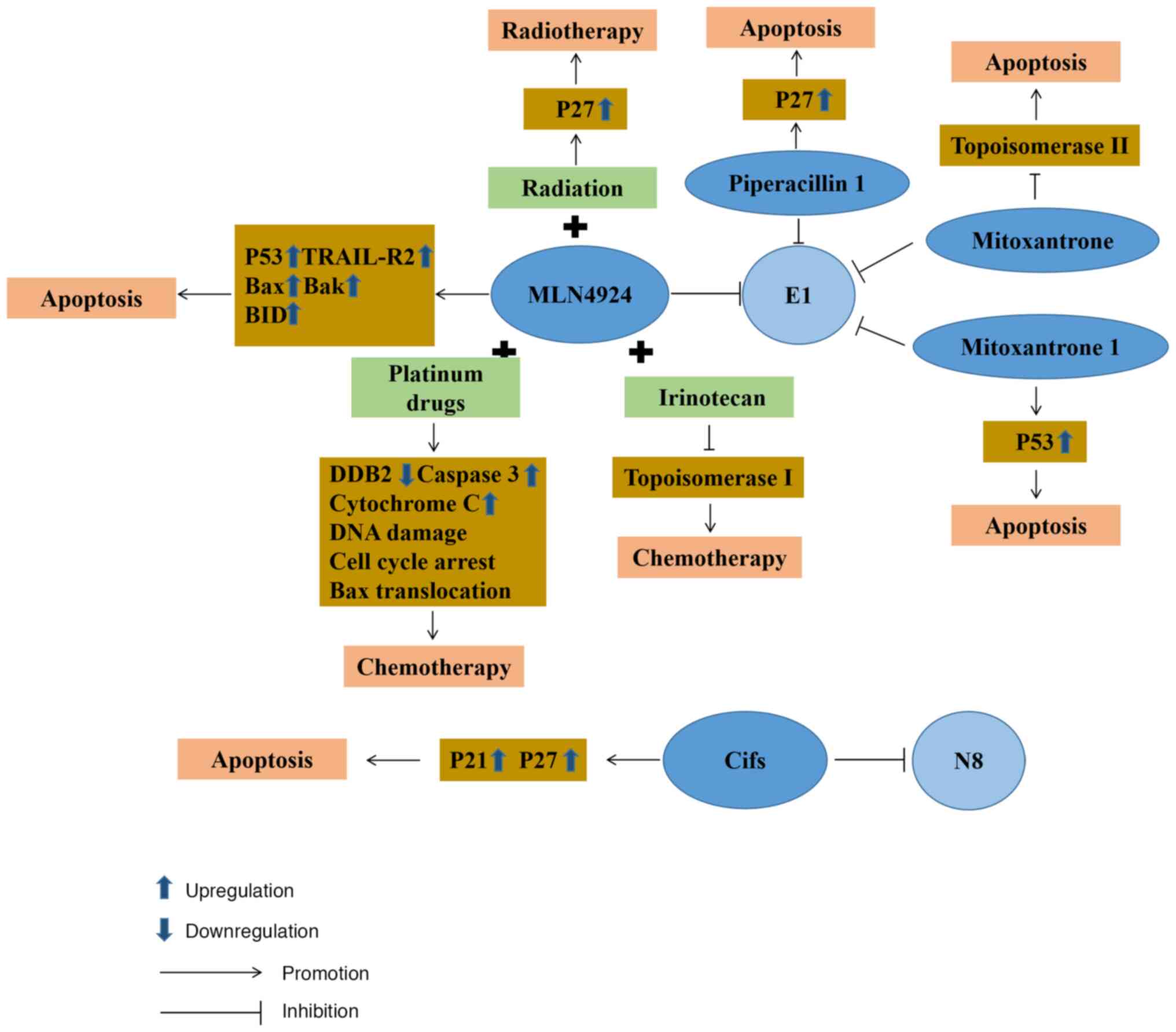
MLN4924
Neddylation plays a crucial role in the progression of CRC. Therefore, the process of developing pharmaceutical interventions targeting NEDD8 has prompted notable advancements in the treatment of CRC. MLN4924 is a selective inhibitor of NAE1. It was first confirmed that MLN4924 could affect the activity of E1 to disrupt the cullin-ring ligase-mediated protein turnover as a NEDD8 substrate (124). Subsequently, further research demonstrated that MLN4924 functions as a selective inhibitor by competitively inhibiting the assembly of the NEDD8-MLN4924 compound. When it is deprived of its capacity to partake in subsequent enzymatic reactions, MLN4924 blocks neddylation in CRC cells and ultimately ushers in the onset of cellular death (125). As research on MLN4924 progressed further, its role as a radiosensitizer was demonstrated, illustrating that it could make CRC cells more sensitive to radiation, primarily through p27 accumulation in cells significantly improving the arrest of radiation-induced cell cycle G2/M phases, DNA damage responses and cellular death (53). The method of CRC cell death using MLN4924 was subsequently further studied. MLN4924 stabilizes p53 to induce ribosome stress, leading to the death of tumor cells. Mitoxantrone 1 also has similar action characteristics (126). As a typical p53 target gene death receptor, TRAIL receptor 2 expression is also significantly upregulated during the process (127). On the other hand, MLN4924 induces cell death by activating Bax and Bak on the outer mitochondrial membrane to initiate mitochondrial outer membrane permeabilization (127). During this biological process, there is also the upregulation in the expression of BH3 interacting domain death agonist (BID). As a member of the pro-apoptotic Bcl2 family, BID activates the downstream targets of caspase-8 in the exogenous pathway (128), causing CRC tumor cell death. Currently, the application of MLN4924 in the treatment of CRC is still being studied in the phase I/II clinical stages. These experimental results demonstrate that this drug can control the progression of cancer through the apoptosis of cancer cells (129,130). MLN4924 is additionally being used in synergy with conventional chemotherapy agents to enhance their efficacy significantly. For example, MLN4924 can induce DNA damage response and upregulate cell cycle checkpoint kinase 2 protein expression levels to render CRC cells more sensitive to oxaliplatin (131). At the same time, MLN4924 can also be combined with irinotecan as a chemotherapeutic method to combat chemotherapy-refractory p53 mutant CRC (127).
However, with the use of MLN4924, CRC tissues will inevitably become resistant. This may be related to FLICE inhibitory protein in MLN4924-treated cells, which leads to resistance to standard care therapies (127). It may also be associated with the variation of NAE, which reduces the affinity of the enzyme to MLN4924 and ATP and increases NEDD8 activation (132).
Beyond its anticipated therapeutic effects in targeted therapy, MLN4924 has demonstrated certain unintended consequences that may foster cancer cell proliferation. These outcomes are primarily associated with the activation of intracellular ERK and JNK signaling, leading to activator protein-1 activation (133). Simultaneously, the inhibitory effects of MLN4924-induced neddylation suppression can upregulate T-cell negative regulator programmed death-ligand 1 (PD-L1) expression (134). Cancer cells exhibiting a high expression of PD-L1 can attenuate T-cell cytotoxicity through the activation of the PD-1/PD-L1 axis, inducing immune suppression (135). Combining anti-PD-L1 antibodies or mitogen-activated protein kinase inhibitors with MLN4924 administration offers a potential avenue to address these challenges effectively (133).
Drug repurposing to screen E1 inhibitors
Since developing new drugs remains costly, there is a trend to seek to repurpose existing approved and investigational drugs given their known safety profile and to reduce costs (136). Piperacillin, known as a β-lactam antibiotic, is often used for the management of suspected bacterial infections relying on empirical treatment combined with tazobactam (137). It is frequently used in cancer patients to treat fever attacks following chemotherapy and neutropenia (138,139). Based on the integrated virtual screening method, it has been found that piperacillin 1 (140) can inhibit the degradation of p27, known as a downstream protein substrate of NAE1, the regulatory subunit of E1. It possesses an ATP-competitive inhibitor without the ability to form the covalent bonds of E1 in the future. Mitoxantrone, as a DNA topoisomerase II poison, causes tumor cell apoptosis by inhibiting DNA synthesis and delaying cell cycle progression, which can improve the therapeutic properties of anthracyclines (141,142). Through an FDA-approved drug database analysis by virtual screening, mitoxantrone 1 (126) was found to have highly selective NAE activity and to compete with ATP, with great potential as an inhibitor of E1.
Cycle inhibitory factors (Cifs)
Cifs are capable of impeding the activity of cullin-ring E3 ubiquitin ligases, resulting in cell cycle arrest. At the same time, Cifs also participate in cancer progression by deamidating NEDD8 (143). In CRC cells, cells that regulate Cif expression by doxycycline lead to tumor apoptosis through p21 and p27 accumulation (144). In terms of specific use, attenuated Salmonella typhimurium VNP20009 can be used to deliver Cif genes to tumor cells, inducing the expression and intracellular accumulation of proteins p27 and p21, thereby inhibiting the growth of tumor cells (145).
Platinum drugs
Platinum-based pharmaceuticals can be employed in the management of CRC and are amenable to concomitant administration with various classes of antineoplastic agents. Commonly used platinum drugs include oxaliplatin, carboplatin and cisplatin, all of which have received global approval (146). Cisplatin is a first-generation platinum drug and interferes with DNA repair by crosslinking with the purine bases of DNA, leading to DNA damage and stimulating tumor cell death (147). Nevertheless, cisplatin lacks the specificity to selectively target CRC cells, consequently leading to diminished accumulative concentrations and a discernible impact on the therapeutic efficacy (148). During the analysis of tissues from patients with squamous cell carcinoma in the head and neck, it was discovered that MLN4924 possesses the capacity to impede the transcription of DDB2 facilitated by E2F1 (149). Since DDB2 demonstrates a crucial function in modulating sensitivity to cisplatin, the combined application of MLN4924 and cisplatin can increase the activity of cisplatin. Moreover, tissues with a suppressed expression of cullin4A exhibit higher susceptibility to cisplatin, which potentially relates to its connection with DNA repair. In addition, the enhancing effect of MLN4924 on cisplatin has been found in breast cancer (150), pancreatic cancer (151) and cervical tumors (152). It can be concluded that the combined application of MLN4924 and cisplatin in CRC tissues may also enhance the sensitivity of cisplatin to compensate for the impact of its low accumulation concentration. Carboplatin is a cisplatin derivative and a second-generation platinum drug whose mode of action is similar to that of cisplatin. It induces tumor cell apoptosis by damaging DNA. Compared with cisplatin, carboplatin produces inter-chain and intra-chain doublets or single adducts following application (153). In individuals with advanced solid tumors, a clinical phase I trial revealed that E1 inhibitor, MLN4924, combined with carboplatin was well-tolerated and stood as a promising benchmark for forthcoming drug development against CRC (130).
Oxaliplatin, a third-generation platinum therapeutic agent, is a first-line chemotherapy drug. As one of the more commonly used chemotherapeutic drugs following surgical resection, it plays a pivotal role in the management of CRC. It initiates the death of malignant cells by orchestrating a halt in cell cycle progression at the critical juncture of G2/M, thus initiating an apoptotic cascade that includes Bax mitochondrial translocation, the release of cytochrome c and caspase-3 catalytic activation (154). In a previous a study on CRC tissues, it was found that MLN4924 combined with oxaliplatin increased oxaliplatin-induced apoptosis (131). Although it has not yet entered clinical trials for CRC, phase I trials have been carried out in the therapeutic management of gastric carcinoma (155). CRC cells have also developed some drug resistance to platinum agents, such as the inhibition of drug accumulation in tumor cells, as well as the acquisition of EMT. A large number of studies on drug resistance have found that exosomes loaded with miR-128-3p (156) and the application of nanoparticles (157) can re-sensitize CRC cells to platinum drugs in vivo.
Irinotecan
Irinotecan is used as a second-line chemotherapeutic agent employed in the treatment of patients with advanced stages of CRC in the event that first-line chemotherapy drug treatment fails. It not only selectively inhibits topoisomerase I, causing tumor cell death by inhibiting DNA function, but also has potent anticancer functions following intracellular modification in cells (158). SN38, the metabolite of irinotecan in vivo, has a synergistic effect with MLN4924. Apoptosis induced by this pathway proceeds in a manner that does not depend on the presence of p53 and circumvents the effects of TP53 mutations in advanced-stag CRC cells (127). This reveals a novel pathway for managing patients grappling with advanced CRC, particularly those battling chemotherapy-resistant p53 mutant CRC. However, it has not yet entered the clinical trial stage. In terms of drug resistance, epigenetic alterations are likely to cause resistance to irinotecan, such as the acetylation of histones in CRC. Therefore, partially drug-resistant CRC can be combated by combining histone deacetylase inhibitors with irinotecan (159) (Fig. 3).
5. Conclusion and future perspectives
In recent years, post-translational modifications of proteins have gradually attracted wide attention. Owing to this, a more in-depth understanding of the ubiquitin-like modification of neddylation has been obtained. Existing research evidence indicates that NEDD8 is primarily related to the proliferation, migration, survival and genetic alterations of tumor cells, and to the microenvironment of tumorigenesis. In CRC, NEDD8 is of paramount significance in the onset and progression of the disease, particularly within the realms of tumor cell migration, proliferation and viability. It is through these mechanisms that anticancer pharmaceuticals directed toward NEDD8 offer novel insight into therapeutic approaches for CRC. They mainly target NAEs for anticancer treatment. There are also drugs, such as cycle inhibitors that target the NEDD8 substrate cullin-ring E3 ubiquitin ligase for anticancer effects. Simultaneously, amalgamated pharmacological interventions, exemplified by the synergistic deployment of MLN4924 alongside irinotecan or by the combination of MLN4924 and oxaliplatin, represent promising strategies in the battle against partially resistant or recalcitrant CRC.
CRC poses one of the greatest threats to human life and health worldwide. Consequently, relentless exploration into the etiology and therapeutic modalities for CRC is imperative. As NEDD8 emerges as a nascent frontier in the arena of anticancer targets, it necessitates a more profound investigation. At present, treatments mainly target NAEs. Regrettably, limited attention has been devoted to agents targeting E2, E3 and NEDD8 substrates of the NEDD8 pathway. The potential emergence of NEDD8 substrates, including Smurf, IκB, cullin 5 and PTEN, as novel targets for therapeutic intervention, holds promise. Simultaneously, the deneddylation of CSN5 exhibits pharmaceutical design potential. Henceforth, other more effective targeted drugs need to be designed for the pathways that cause CRC. At the same time, for some NEDD8 target drugs that have been used, drug resistance issues should also be paid investigated during the period of treatment, such as the emergence of MLN4924 resistance. In future research, an exploration of novel therapeutic analogs or strategies to combat drug resistance is paramount. Consequently, studying the pathogenesis of CRC based on NEDD8 and unearthing drug targets for CRC are of utmost importance for the prognosis and survival of patients with CRC.
Availability of data and materials
Not applicable.
Authors' contributions
TW and XL contributed equally to the present study; they were involved in the conception and design of the study, and in the drafting of the manuscript. RM and JS were involved in the analysis of the literature. SH, ZS and MW conceived and supervised the study, and directed the writing. All the authors have read and approved the final version of this review. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Abbreviations:
|
Bcl-2 |
B-cell lymphoma 2 |
|
Cifs |
cycle inhibitory factors |
|
CRC |
colorectal cancer |
|
CRL |
cullin-ring ligase |
|
CSN |
COP9 signalosome |
|
EMT |
epithelial-to-mesenchymal transition |
|
ERK |
extracellular signal-regulated kinase |
|
FASN |
fatty acid synthase |
|
HIF |
hypoxia-inducible factor |
|
HuR |
Hu protein R |
|
MDM2 |
murine double minute 2 |
|
MSI |
microsatellite instability |
|
NAE |
NEDD activating enzyme |
|
NEDD8 |
neural precursor cell expressed developmentally downregulated 8 |
|
PCNA |
proliferating cell nuclear antigen |
|
PTEN |
phosphatase and tensin homolog |
|
pVHL |
Von Hippel-Lindau tumor suppressor protein |
|
RhoA |
rat sarcoma homolog gene family A |
|
RRP9 |
ribosomal RNA processing 9 |
|
Skp2 |
s-phase kinase associated protein 2 |
|
Smurf |
SMAD ubiquitination regulatory factor |
|
UBC12 |
ubiquitin-conjugating enzyme C12 |
|
UBE |
ubiquitin-conjugating enzyme |
|
XIAP |
X-linked inhibitor of apoptosis protein |
Acknowledgments
Figures of this review were created with Servier Medical Art (https://smart.servier.com/).
Funding
The present study was supported by the Shandong Natural Science Foundation (grant no. ZR2021QH046).
References
|
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Brenner H, Kloor M and Pox CP: Colorectal cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar | |
|
Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Roque-Castellano C, Fariña-Castro R, Nogués-Ramia EM, Artiles-Armas M and Marchena-Gómez J: Colorectal cancer surgery in selected nonagenarians is relatively safe and it is associated with a good long-term survival: An observational study. World J Surg Oncol. 18:1202020. View Article : Google Scholar : PubMed/NCBI | |
|
Salibasic M, Pusina S, Bicakcic E, Pasic A, Gavric I, Kulovic E, Rovcanin A and Beslija S: Colorectal cancer surgical treatment, our experience. Med Arch. 73:412–414. 2019. View Article : Google Scholar | |
|
Luo W, Wu M and Chen Y: Laparoscopic versus open surgery for elderly patients with colorectal cancer: A systematic review and meta-analysis of matched studies. ANZ J Surg. 92:2003–2017. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Ustuner MA, Deniz A and Simsek A: Laparoscopic <em>versus</em> open surgery in colorectal cancer: Is laparoscopy safe enough? J Coll Physicians Surg Pak. 32:1170–1174. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Liu G, Zhang S, Zhang Y, Fu X and Liu X: Robotic surgery in rectal cancer: Potential, challenges, and opportunities. Curr Treat Options Oncol. 23:961–979. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Riesco-Martinez MC, Modrego A, Espinosa-Olarte P, La Salvia A and Garcia-Carbonero R: Perioperative chemotherapy for liver metastasis of colorectal cancer: Lessons learned and future perspectives. Curr Treat Options Oncol. 23:1320–1337. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Habr-Gama A, Perez RO, São Julião GP, Proscurshim I and Gama-Rodrigues J: Nonoperative approaches to rectal cancer: A critical evaluation. Semin Radiat Oncol. 21:234–239. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Hsu YJ, Chern YJ, Lai IL, Chiang SF, Liao CK, Tsai WS, Hung HY, Hsieh PS, Yeh CY, Chiang JM, et al: Usefulness of close surveillance for rectal cancer patients after neoadjuvant chemoradiotherapy. Open Med (Wars). 17:1438–1448. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
McWhirter D, Kitteringham N, Jones RP, Malik H, Park K and Palmer D: Chemotherapy induced hepatotoxicity in metastatic colorectal cancer: A review of mechanisms and outcomes. Crit Rev Oncol Hematol. 88:404–415. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Kim JH: Chemotherapy for colorectal cancer in the elderly. World J Gastroenterol. 21:5158–5166. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Piawah S and Venook AP: Targeted therapy for colorectal cancer metastases: A review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer. 125:4139–4147. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Kumar S, Tomooka Y and Noda M: Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 185:1155–1161. 1992. View Article : Google Scholar : PubMed/NCBI | |
|
Kumar S, Yoshida Y and Noda M: Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem Biophys Res Commun. 195:393–399. 1993. View Article : Google Scholar : PubMed/NCBI | |
|
Kamitani T, Kito K, Nguyen HP and Yeh ET: Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 272:28557–28562. 1997. View Article : Google Scholar : PubMed/NCBI | |
|
Wada H, Kito K, Caskey LS, Yeh ET and Kamitani T: Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem Biophys Res Commun. 251:688–692. 1998. View Article : Google Scholar : PubMed/NCBI | |
|
Mendoza HM, Shen LN, Botting C, Lewis A, Chen J, Ink B and Hay RT: NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem. 278:25637–25643. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Gong L and Yeh ET: Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 274:12036–12042. 1999. View Article : Google Scholar : PubMed/NCBI | |
|
Huang DT, Zhuang M, Ayrault O and Schulman BA: Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nat Struct Mol Biol. 15:280–287. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF and Schulman BA: E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 33:483–495. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Baek K, Scott DC and Schulman BA: NEDD8 and ubiquitin ligation by cullin-RING E3 ligases. Curr Opin Struct Biol. 67:101–109. 2021. View Article : Google Scholar : | |
|
Lydeard JR, Schulman BA and Harper JW: Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 14:1050–1061. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Xirodimas DP, Saville MK, Bourdon JC, Hay RT and Lane DP: Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 118:83–97. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Stickle NH, Chung J, Klco JM, Hill RP, Kaelin WG Jr and Ohh M: pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol. 24:3251–3261. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Guan J, Yu S and Zheng X: NEDDylation antagonizes ubiquitination of proliferating cell nuclear antigen and regulates the recruitment of polymerase η in response to oxidative DNA damage. Protein Cell. 9:365–379. 2018. | |
|
Ryu JH, Li SH, Park HS, Park JW, Lee B and Chun YS: Hypoxia-inducible factor α subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. J Biol Chem. 286:6963–6970. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Brown JS and Jackson SP: Ubiquitylation, neddylation and the DNA damage response. Open Biol. 5:1500182015. View Article : Google Scholar : PubMed/NCBI | |
|
Liu J and Nussinov R: Flexible cullins in cullin-RING E3 ligases allosterically regulate ubiquitination. J Biol Chem. 286:40934–40942. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Sarikas A, Hartmann T and Pan ZQ: The cullin protein family. Genome Biol. 12:2202011. View Article : Google Scholar : PubMed/NCBI | |
|
Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K and Kato S: A new NEDD8-ligating system for cullin-4A. Genes Dev. 12:2263–2268. 1998. View Article : Google Scholar : PubMed/NCBI | |
|
Petroski MD and Deshaies RJ: Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 6:9–20. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Nam SY, Ko YS, Jung J, Yoon J, Kim YH, Choi YJ, Park JW, Chang MS, Kim WH and Lee BL: A hypoxia-dependent upregulation of hypoxia-inducible factor-1 by nuclear factor-κB promotes gastric tumour growth and angiogenesis. Br J Cancer. 104:166–174. 2011. View Article : Google Scholar | |
|
Semenza GL: HIF-1 and mechanisms of hypoxia sensing. Curr Opin Cell Biol. 13:167–171. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Hanahan D and Weinberg RA: Hallmarks of cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Kittai AS, Danilova OV, Lam V, Liu T, Bruss N, Best S, Fan G and Danilov AV: NEDD8-activating enzyme inhibition induces cell cycle arrest and anaphase catastrophe in malignant T-cells. Oncotarget. 12:2068–2074. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Lan H, Tang Z, Jin H and Sun Y: Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells. Sci Rep. 6:242182016. View Article : Google Scholar : PubMed/NCBI | |
|
Liu H, Bei Q and Luo X: MLN4924 inhibits cell proliferation by targeting the activated neddylation pathway in endometrial carcinoma. J Int Med Res. 49:30006052110185922021.PubMed/NCBI | |
|
Picco G, Petti C, Sassi F, Grillone K, Migliardi G, Rossi T, Isella C, Di Nicolantonio F, Sarotto I, Sapino A, et al: Efficacy of NEDD8 pathway inhibition in preclinical models of poorly differentiated, clinically aggressive colorectal cancer. J Natl Cancer Inst. 109:djw2092017. View Article : Google Scholar | |
|
Xie P, Zhang M, He S, Lu K, Chen Y, Xing G, Lu Y, Liu P, Li Y, Wang S, et al: The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nat Commun. 5:37332014. View Article : Google Scholar : PubMed/NCBI | |
|
Jiang Y, Liang Y, Li L, Zhou L, Cheng W, Yang X, Yang X, Qi H, Yu J, Jeong LS, et al: Targeting neddylation inhibits intravascular survival and extravasation of cancer cells to prevent lung-cancer metastasis. Cell Biol Toxicol. 35:233–245. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Mickova A, Kharaishvili G, Kurfurstova D, Gachechiladze M, Kral M, Vacek O, Pokryvkova B, Mistrik M, Soucek K and Bouchal J: Skp2 and slug are coexpressed in aggressive prostate cancer and inhibited by neddylation blockade. Int J Mol Sci. 22:28442021. View Article : Google Scholar : PubMed/NCBI | |
|
Tan KL and Pezzella F: Inhibition of NEDD8 and FAT10 ligase activities through the degrading enzyme NEDD8 ultimate buster 1: A potential anticancer approach. Oncol Lett. 12:4287–4296. 2016. View Article : Google Scholar | |
|
Watson IR, Blanch A, Lin DC, Ohh M and Irwin MS: Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J Biol Chem. 281:34096–34103. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Aoki I, Higuchi M and Gotoh Y: NEDDylation controls the target specificity of E2F1 and apoptosis induction. Oncogene. 32:3954–3964. 2013. View Article : Google Scholar | |
|
Halazonetis TD, Gorgoulis VG and Bartek J: An oncogene-induced DNA damage model for cancer development. Science. 319:1352–1355. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Garvin AJ: Beyond reversal: ubiquitin and ubiquitin-like proteases and the orchestration of the DNA double strand break repair response. Biochem Soc Trans. 47:1881–1893. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Meir M, Galanty Y, Kashani L, Blank M, Khosravi R, Fernández-Ávila MJ, Cruz-García A, Star A, Shochot L, Thomas Y, et al: The COP9 signalosome is vital for timely repair of DNA double-strand breaks. Nucleic Acids Res. 43:4517–4530. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Gâtel P, Piechaczyk M and Bossis G: Ubiquitin, SUMO, and Nedd8 as therapeutic targets in cancer. Adv Exp Med Biol. 1233:29–54. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye WC, Zhang DM and Chen ZS: Autophagy and multidrug resistance in cancer. Chin J Cancer. 36:522017. View Article : Google Scholar : PubMed/NCBI | |
|
Sun Y, Baechler SA, Zhang X, Kumar S, Factor VM, Arakawa Y, Chau CH, Okamoto K, Parikh A, Walker B, et al: Targeting neddylation sensitizes colorectal cancer to topoisomerase I inhibitors by inactivating the DCAF13-CRL4 ubiquitin ligase complex. Nat Commun. 14:37622023. View Article : Google Scholar : PubMed/NCBI | |
|
Wan J, Zhu J, Li G and Zhang Z: Radiosensitization of human colorectal cancer cells by MLN4924: An Inhibitor of NEDD8-Activating Enzyme. Technol Cancer Res Treat. 15:527–534. 2016. View Article : Google Scholar | |
|
Shao Y, Liu Z, Song X, Sun R, Zhou Y, Zhang D, Sun H, Huang J, Wu C, Gu W, et al: ALKBH5/YTHDF2-mediated m6A modification of circAFF2 enhances radiosensitivity of colorectal cancer by inhibiting Cullin neddylation. Clin Transl Med. 13:e13182023. View Article : Google Scholar : PubMed/NCBI | |
|
Whiteside TL: The tumor microenvironment and its role in promoting tumor growth. Oncogene. 27:5904–5912. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou L, Jiang Y, Luo Q, Li L and Jia L: Neddylation: A novel modulator of the tumor microenvironment. Mol Cancer. 18:772019. View Article : Google Scholar : PubMed/NCBI | |
|
Chang FM, Reyna SM, Granados JC, Wei SJ, Innis-Whitehouse W, Maffi SK, Rodriguez E, Slaga TJ and Short JD: Inhibition of neddylation represses lipopolysaccharide-induced proinflammatory cytokine production in macrophage cells. J Biol Chem. 287:35756–35767. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Li L, Liu B, Dong T, Lee HW, Yu J, Zheng Y, Gao H, Zhang Y, Chu Y, Liu G, et al: Neddylation pathway regulates the proliferation and survival of macrophages. Biochem Biophys Res Commun. 432:494–498. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou L, Jiang Y, Liu X, Li L, Yang X, Dong C, Liu X, Lin Y, Li Y, Yu J, et al: Promotion of tumor-associated macrophages infiltration by elevated neddylation pathway via NF-κB-CCL2 signaling in lung cancer. Oncogene. 38:5792–5804. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Joyce JA and Fearon DT: T cell exclusion, immune privilege, and the tumor microenvironment. Science. 348:74–80. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Jin HS, Liao L, Park Y and Liu YC: Neddylation pathway regulates T-cell function by targeting an adaptor protein Shc and a protein kinase Erk signaling. Proc Natl Acad Sci USA. 110:624–629. 2013. View Article : Google Scholar : | |
|
Jiang Y, Li L, Li Y, Liu G, Hoffman RM and Jia L: Neddylation regulates macrophages and implications for cancer therapy. Front Cell Dev Biol. 9:6811862021. View Article : Google Scholar : PubMed/NCBI | |
|
Best S, Lam V, Liu T, Bruss N, Kittai A, Danilova OV, Murray S, Berger A, Pennock ND, Lind EF and Danilov AV: Immunomodulatory effects of pevonedistat, a NEDD8-activating enzyme inhibitor, in chronic lymphocytic leukemia-derived T cells. Leukemia. 35:156–168. 2021. View Article : Google Scholar : | |
|
Maishi N and Hida K: Tumor endothelial cells accelerate tumor metastasis. Cancer Sci. 108:1921–1926. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Tang N, Wang L, Esko J, Giordano FJ, Huang Y, Gerber HP, Ferrara N and Johnson RS: Loss of HIF-1alpha in endothelial cells disrupts a hypoxia-driven VEGF autocrine loop necessary for tumorigenesis. Cancer Cell. 6:485–495. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Shi CS, Kuo KL, Lin WC, Chen MS, Liu SH, Liao SM, Hsu CH, Chang YW, Chang HC and Huang KH: Neddylation inhibitor, MLN4924 suppresses angiogenesis in huvecs and solid cancers: in vitro and in vivo study. Am J Cancer Res. 10:953–964. 2020.PubMed/NCBI | |
|
Yao WT, Wu JF, Yu GY, Wang R, Wang K, Li LH, Chen P, Jiang YN, Cheng H, Lee HW, et al: Suppression of tumor angiogenesis by targeting the protein neddylation pathway. Cell Death Dis. 5:e10592014. View Article : Google Scholar : PubMed/NCBI | |
|
Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M, Ibiza S, Cañellas A, Hernando-Momblona X, et al: TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 554:538–543. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Kerscher O, Felberbaum R and Hochstrasser M: Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 22:159–180. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
He S, Cao Y, Xie P, Dong G and Zhang L: The Nedd8 Non-covalent Binding Region in the Smurf HECT Domain is Critical to its Ubiquitn Ligase Function. Sci Rep. 7:413642017. View Article : Google Scholar : PubMed/NCBI | |
|
Zheng J, Shi Z, Yang P, Zhao Y, Tang W, Ye S, Xuan Z, Chen C, Shao C, Wu Q and Sun H: ERK-Smurf1-RhoA signaling is critical for TGFβ-drived EMT and tumor metastasis. Life Sci Alliance. 5:e2021013302022. View Article : Google Scholar | |
|
Xie L, Law BK, Chytil AM, Brown KA, Aakre ME and Moses HL: Activation of the Erk pathway is required for TGF-beta1-induced EMT in vitro. Neoplasia. 6:603–610. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Fang JY and Richardson BC: The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 6:322–327. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Cardoso AP, Pinto ML, Pinto AT, Oliveira MI, Pinto MT, Gonçalves R, Relvas JB, Figueiredo C, Seruca R, Mantovani A, et al: Macrophages stimulate gastric and colorectal cancer invasion through EGFR Y(1086), c-Src, Erk1/2 and Akt phosphorylation and smallGTPase activity. Oncogene. 33:2123–2133. 2014. View Article : Google Scholar | |
|
Price JT, Wilson HM and Haites NE: Epidermal growth factor (EGF) increases the in vitro invasion, motility and adhesion interactions of the primary renal carcinoma cell line, A704. Eur J Cancer. 32A:1977–1982. 1996. View Article : Google Scholar : PubMed/NCBI | |
|
Li S, Wu X, Xu Y, Wu S, Li Z, Chen R, Huang N, Zhu Z and Xu X: miR-145 suppresses colorectal cancer cell migration and invasion by targeting an ETS-related gene. Oncol Rep. 36:1917–1926. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Kwon A, Lee HL, Woo KM, Ryoo HM and Baek JH: SMURF1 plays a role in EGF-induced breast cancer cell migration and invasion. Mol Cells. 36:548–555. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT and Horwitz AR: Cell migration: Integrating signals from front to back. Science. 302:1704–1709. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Du MG, Liu F, Chang Y, Tong S, Liu W, Chen YJ and Xie P: Neddylation modification of the U3 snoRNA-binding protein RRP9 by Smurf1 promotes tumorigenesis. J Biol Chem. 297:1013072021. View Article : Google Scholar : PubMed/NCBI | |
|
Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, et al: High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 307:1621–1625. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Clerget G, Bourguignon-Igel V, Marmier-Gourrier N, Rolland N, Wacheul L, Manival X, Charron C, Kufel J, Méreau A, Senty-Ségault V, et al: Synergistic defects in pre-rRNA processing from mutations in the U3-specific protein Rrp9 and U3 snoRNA. Nucleic Acids Res. 48:3848–3868. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Pecoraro A, Pagano M, Russo G and Russo A: Ribosome Biogenesis and Cancer: Overview on Ribosomal Proteins. Int J Mol Sci. 22:54962021. View Article : Google Scholar : PubMed/NCBI | |
|
Pelletier J, Thomas G and Volarević S: Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat Rev Cancer. 18:51–63. 2018. View Article : Google Scholar | |
|
Guo J, Xu G, Mao C and Wei R: Low Expression of Smurf1 Enhances the Chemosensitivity of Human Colorectal Cancer to Gemcitabine and Cisplatin in Patient-Derived Xenograft Models. Transl Oncol. 13:1008042020. View Article : Google Scholar : PubMed/NCBI | |
|
Song MS, Salmena L and Pandolfi PP: The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 13:283–296. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Salmena L, Carracedo A and Pandolfi PP: Tenets of PTEN tumor suppression. Cell. 133:403–414. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Xie P, Peng Z, Chen Y, Li H, Du M, Tan Y, Zhang X, Lu Z, Cui CP, Liu CH, et al: Neddylation of PTEN regulates its nuclear import and promotes tumor development. Cell Res. 31:291–311. 2021. View Article : Google Scholar : | |
|
Finicle BT, Jayashankar V and Edinger AL: Nutrient scavenging in cancer. Nat Rev Cancer. 18:619–633. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Murphy N, Song M, Papadimitriou N, Carreras-Torres R, Langenberg C, Martin RM, Tsilidis KK, Barroso I, Chen J, Frayling TM, et al: Associations Between Glycemic Traits and Colorectal Cancer: A Mendelian Randomization Analysis. J Natl Cancer Inst. 114:740–752. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang W, Lu Y and Li X, Zhang J, Lin W, Zhang W, Zheng L and Li X: IPO5 promotes the proliferation and tumourigenicity of colorectal cancer cells by mediating RASAL2 nuclear transportation. J Exp Clin Cancer Res. 38:2962019. View Article : Google Scholar : PubMed/NCBI | |
|
Menendez JA and Lupu R: Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 7:763–777. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Xia Q, Zhang H, Zhang P, Li Y, Xu M, Li X, Li X and Dong L: Oncogenic Smurf1 promotes PTEN wild-type glioblastoma growth by mediating PTEN ubiquitylation. Oncogene. 39:5902–5915. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Du MG, Peng ZQ, Gai WB, Liu F, Liu W, Chen YJ, Li HC, Zhang X, Liu CH, Zhang LQ, et al: The Absence of PTEN in Breast Cancer Is a Driver of MLN4924 Resistance. Front Cell Dev Biol. 9:6674352021. View Article : Google Scholar : PubMed/NCBI | |
|
Phelps RA, Chidester S, Dehghanizadeh S, Phelps J, Sandoval IT, Rai K, Broadbent T, Sarkar S, Burt RW and Jones DA: A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 137:623–634. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Schütz AK, Hennes T, Jumpertz S, Fuchs S and Bernhagen J: Role of CSN5/JAB1 in Wnt/β-catenin activation in colorectal cancer cells. FEBS Lett. 586:1645–1651. 2012. View Article : Google Scholar | |
|
Cope GA and Deshaies RJ: COP9 signalosome: A multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 114:663–671. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Jumpertz S, Hennes T, Asare Y, Vervoorts J, Bernhagen J and Schütz AK: The β-catenin E3 ubiquitin ligase SIAH-1 is regulated by CSN5/JAB1 in CRC cells. Cell Signal. 26:2051–2059. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Sninsky JA, Shore BM, Lupu GV and Crockett SD: Risk Factors for Colorectal Polyps and Cancer. Gastrointest Endosc Clin N Am. 32:195–213. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Aiello NM, Maddipati R, Norgard RJ, Balli D, Li J, Yuan S, Yamazoe T, Black T, Sahmoud A, Furth EE, et al: EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration. Dev Cell. 45:681–695.e84. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Asmamaw MD, Liu Y, Zheng YC, Shi XJ and Liu HM: Skp2 in the ubiquitin-proteasome system: A comprehensive review. Med Res Rev. 40:1920–1949. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Serrano-Gomez SJ, Maziveyi M and Alahari SK: Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer. 15:182016. View Article : Google Scholar : PubMed/NCBI | |
|
Yu X, Zhou L, Liu W, Liu L, Gao F, Li W and Liu H: Skp2 stabilizes Mcl-1 and confers radioresistance in colorectal cancer. Cell Death Dis. 13:2492022. View Article : Google Scholar : PubMed/NCBI | |
|
Chen P, Li X, Zhang R, Liu S, Xiang Y, Zhang M, Chen X, Pan T, Yan L, Feng J, et al: Combinative treatment of β-elemene and cetuximab is sensitive to KRAS mutant colorectal cancer cells by inducing ferroptosis and inhibiting epithelial-mesenchymal transformation. Theranostics. 10:5107–5119. 2020. View Article : Google Scholar : | |
|
Wang L, Li S, Luo H, Lu Q and Yu S: PCSK9 promotes the progression and metastasis of colon cancer cells through regulation of EMT and PI3K/AKT signaling in tumor cells and phenotypic polarization of macrophages. J Exp Clin Cancer Res. 41:3032022. View Article : Google Scholar : PubMed/NCBI | |
|
Kunkel TA and Erie DA: DNA mismatch repair. Annu Rev Biochem. 74:681–710. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Guastadisegni C, Colafranceschi M, Ottini L and Dogliotti E: Microsatellite instability as a marker of prognosis and response to therapy: A meta-analysis of colorectal cancer survival data. Eur J Cancer. 46:2788–2798. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
de la Chapelle A and Hampel H: Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol. 28:3380–3387. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
McGrail DJ, Garnett J, Yin J, Dai H, Shih DJH, Lam TNA, Li Y, Sun C, Li Y, Schmandt R, et al: Proteome Instability Is a Therapeutic Vulnerability in Mismatch Repair-Deficient Cancer. Cancer Cell. 37:371–386.e12. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Johnstone RW, Ruefli AA and Lowe SW: Apoptosis: A link between cancer genetics and chemotherapy. Cell. 108:153–164. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ and Cheng EH: Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol. 8:1348–1358. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Wong WW and Puthalakath H: Bcl-2 family proteins: The sentinels of the mitochondrial apoptosis pathway. IUBMB Life. 60:390–397. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Ploner C, Kofler R and Villunger A: Noxa: At the tip of the balance between life and death. Oncogene. 27(Suppl 1): S84–S92. 2008. View Article : Google Scholar | |
|
Xu S, Ma Y, Tong Q, Yang J, Liu J, Wang Y, Li G, Zeng J, Fang S, Li F, et al: Cullin-5 neddylation-mediated NOXA degradation is enhanced by PRDX1 oligomers in colorectal cancer. Cell Death Dis. 12:2652021. View Article : Google Scholar : PubMed/NCBI | |
|
Brennan CM and Steitz JA: HuR and mRNA stability. Cell Mol Life Sci. 58:266–277. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Wang W, Caldwell MC, Lin S, Furneaux H and Gorospe M: HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 19:2340–2350. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Abdelmohsen K and Gorospe M: Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA. 1:214–229. 2010. View Article : Google Scholar | |
|
McLarnon A: Cancer: Mdm2-regulated stabilization of HuR by neddylation in HCC and colon cancer-a possible target for therapy. Nat Rev Gastroenterol Hepatol. 9:42011. | |
|
Embade N, Fernández-Ramos D, Varela-Rey M, Beraza N, Sini M, Gutiérrez de Juan V, Woodhoo A, Martínez-López N, Rodríguez-Iruretagoyena B, Bustamante FJ, et al: Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology. 55:1237–1248. 2012. View Article : Google Scholar | |
|
Greenlee JD, Lopez-Cavestany M, Ortiz-Otero N, Liu K, Subramanian T, Cagir B and King MR: Oxaliplatin resistance in colorectal cancer enhances TRAIL sensitivity via death receptor 4 upregulation and lipid raft localization. Elife. 10:e677502021. View Article : Google Scholar : PubMed/NCBI | |
|
Lee SJ, Lee DE, Choi SY and Kwon OS: OSMI-1 Enhances TRAIL-Induced Apoptosis through ER Stress and NF-κB Signaling in Colon Cancer Cells. Int J Mol Sci. 22:110732021. View Article : Google Scholar | |
|
Paiva C, Godbersen JC, Rowland T, Danilova OV, Danes C, Berger A and Danilov AV: Pevonedistat, a Nedd8-activating enzyme inhibitor, sensitizes neoplastic B-cells to death receptor-mediated apoptosis. Oncotarget. 8:21128–21139. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Sakamoto K, Maeda S, Hikiba Y, Nakagawa H, Hayakawa Y, Shibata W, Yanai A, Ogura K and Omata M: Constitutive NF-kappaB activation in colorectal carcinoma plays a key role in angiogenesis, promoting tumor growth. Clin Cancer Res. 15:2248–2258. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Linares J, Sallent-Aragay A, Badia-Ramentol J, Recort-Bascuas A, Méndez A, Manero-Rupérez N, Re DL, Rivas EI, Guiu M, Zwick M, et al: Long-term platinum-based drug accumulation in cancer-associated fibroblasts promotes colorectal cancer progression and resistance to therapy. Nat Commun. 14:7462023. View Article : Google Scholar : PubMed/NCBI | |
|
Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al: An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 458:732–736. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, et al: Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: The NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 37:102–111. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Wu KJ, Zhong HJ, Li G, Liu C, Wang HD, Ma DL and Leung CH: Structure-based identification of a NEDD8-activating enzyme inhibitor via drug repurposing. Eur J Med Chem. 143:1021–1027. 2018. View Article : Google Scholar | |
|
Ferris J, Espona-Fiedler M, Hamilton C, Holohan C, Crawford N, McIntyre AJ, Roberts JZ, Wappett M, McDade SS, Longley DB and Coyle V: Pevonedistat (MLN4924): Mechanism of cell death induction and therapeutic potential in colorectal cancer. Cell Death Discov. 6:612020. View Article : Google Scholar : PubMed/NCBI | |
|
Li H, Zhu H, Xu CJ and Yuan J: Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 94:491–501. 1998. View Article : Google Scholar : PubMed/NCBI | |
|
Sekeres MA, Watts J, Radinoff A, Sangerman MA, Cerrano M, Lopez PF, Zeidner JF, Campelo MD, Graux C, Liesveld J, et al: Randomized phase 2 trial of pevonedistat plus azacitidine versus azacitidine for higher-risk MDS/CMML or low-blast AML. Leukemia. 35:2119–2124. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou X, Sedarati F, Faller DV, Zhao D, Faessel HM, Chowdhury S, Bolleddula J, Li Y, Venkatakrishnan K and Papai Z: Phase I study assessing the mass balance, pharmacokinetics, and excretion of [14C]-pevonedistat, a NEDD8-activating enzyme inhibitor in patients with advanced solid tumors. Invest New Drugs. 39:488–498. 2021. View Article : Google Scholar | |
|
Zheng W, Luo Z, Zhang J, Min P, Li W, Xu D, Zhang Z, Xiong P, Liang H and Liu J: Neural precursor cell expressed, developmentally downregulated 8-activating enzyme inhibitor MLN4924 sensitizes colorectal cancer cells to oxaliplatin by inducing DNA damage, G2 cell cycle arrest and apoptosis. Mol Med Rep. 15:2795–2801. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Toth JI, Yang L, Dahl R and Petroski MD: A gatekeeper residue for NEDD8-activating enzyme inhibition by MLN4924. Cell Rep. 1:309–316. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang S, You X, Xu T, Chen Q, Li H, Dou L and Sun Y, Xiong X, Meredith MA and Sun Y: PD-L1 induction via the MEK-JNK-AP1 axis by a neddylation inhibitor promotes cancer-associated immunosuppression. Cell Death Dis. 13:8442022. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou S, Zhao X, Yang Z, Yang R, Chen C, Zhao K, Wang W, Ma Y, Zhang Q and Wang X: Neddylation inhibition upregulates PD-L1 expression and enhances the efficacy of immune checkpoint blockade in glioblastoma. Int J Cancer. 145:763–774. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Gong J, Chehrazi-Raffle A, Reddi S and Salgia R: Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: A comprehensive review of registration trials and future considerations. J Immunother Cancer. 6:82018. View Article : Google Scholar : PubMed/NCBI | |
|
Issa NT, Stathias V, Schürer S and Dakshanamurthy S: Machine and deep learning approaches for cancer drug repurposing. Semin Cancer Biol. 68:132–142. 2021. View Article : Google Scholar | |
|
Gin A, Dilay L, Karlowsky JA, Walkty A, Rubinstein E and Zhanel GG: Piperacillin-tazobactam: A beta-lactam/beta-lactamase inhibitor combination. Expert Rev Anti Infect Ther. 5:365–383. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Maarbjerg SF, Thorsted A, Friberg LE, Nielsen EI, Wang M, Schrøder H and Albertsen BK: Continuous infusion of piperacillin-tazobactam significantly improves target attainment in children with cancer and fever. Cancer Rep (Hoboken). 5:e15852022. View Article : Google Scholar | |
|
Rosanova MT, Cuellar-Pompa L and Lede R: Efficacy and safety of empirical treatment with piperacillin/tazobactan as monotherapy in episodes of neutropenia and fever in children with cancer: Systematic review and meta-analysis. Rev Chilena Infectol. 38:488–494. 2021.In Spanish. View Article : Google Scholar : PubMed/NCBI | |
|
Zhong HJ, Liu LJ, Chan DS, Wang HM, Chan PW, Ma DL and Leung CH: Structure-based repurposing of FDA-approved drugs as inhibitors of NEDD8-activating enzyme. Biochimie. 102:211–215. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Evison BJ, Sleebs BE, Watson KG, Phillips DR and Cutts SM: Mitoxantrone, more than just another topoisomerase II poison. Med Res Rev. 36:248–299. 2016. View Article : Google Scholar | |
|
Faulds D, Balfour JA, Chrisp P and Langtry HD: Mitoxantrone. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in the chemotherapy of cancer. Drugs. 41:400–449. 1991. View Article : Google Scholar : PubMed/NCBI | |
|
Taieb F, Nougayrède JP and Oswald E: Cycle inhibiting factors (cifs): Cyclomodulins that usurp the ubiquitin-dependent degradation pathway of host cells. Toxins (Basel). 3:356–368. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Liu L, Ni J, Zhang J and He X: Construction and characterization of regulated cycle inhibiting factors induced upon Tet-On system in human colon cancer cell lines. Anticancer Drugs. 29:854–860. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Liu L, Zhang J, Gu M, Li G, Ni J and Fan M: Antitumor effect of cycle inhibiting factor expression in colon cancer via salmonella VNP20009. Anticancer Agents Med Chem. 20:1722–1727. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Wheate NJ, Walker S, Craig GE and Oun R: The status of platinum anticancer drugs in the clinic and in clinical trials. Dalton Trans. 39:8113–8127. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Tchounwou PB, Dasari S, Noubissi FK, Ray P and Kumar S: Advances in our understanding of the molecular mechanisms of action of cisplatin in cancer therapy. J Exp Pharmacol. 13:303–328. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Li W, Sun Y, Chen J, Jiang Z and Yang J: PEGylated cisplatin nanoparticles for treating colorectal cancer in a pH-Responsive manner. J Immunol Res. 2022:80239152022. View Article : Google Scholar : PubMed/NCBI | |
|
Jones TM, Espitia CM, Ooi A, Bauman JE, Carew JS and Nawrocki ST: Targeted CUL4A inhibition synergizes with cisplatin to yield long-term survival in models of head and neck squamous cell carcinoma through a DDB2-mediated mechanism. Cell Death Dis. 13:3502022. View Article : Google Scholar : PubMed/NCBI | |
|
Misra S, Zhang X, Wani NA, Sizemore S and Ray A: Both BRCA1-wild type and -mutant triple-negative breast cancers show sensitivity to the NAE inhibitor MLN4924 which is enhanced upon MLN4924 and cisplatin combination treatment. Oncotarget. 11:784–800. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Zeng Y, Iv YS, Pan QH, Zhou YG and Li H: An overactive neddylation pathway serves as a therapeutic target and MLN4924 enhances the anticancer activity of cisplatin in pancreatic cancer. Oncol Lett. 18:2724–2732. 2019.PubMed/NCBI | |
|
Lin WC, Kuo KL, Shi CS, Wu JT, Hsieh JT, Chang HC, Liao SM, Chou CT, Chiang CK, Chiu WS, et al: MLN4924, a Novel NEDD8-activating enzyme inhibitor, exhibits antitumor activity and enhances cisplatin-induced cytotoxicity in human cervical carcinoma: In vitro and in vivo study. Am J Cancer Res. 5:3350–3362. 2015. | |
|
Ho GY, Woodward N and Coward JI: Cisplatin versus carboplatin: Comparative review of therapeutic management in solid malignancies. Crit Rev Oncol Hematol. 102:37–46. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Arango D, Wilson AJ, Shi Q, Corner GA, Arañes MJ, Nicholas C, Lesser M, Mariadason JM and Augenlicht LH: Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. Br J Cancer. 91:1931–1946. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Shoji H, Takahari D, Hara H, Nagashima K, Adachi J and Boku N: A phase I study of pevonedistat plus capecitabine plus oxaliplatin in patients with advanced gastric cancer refractory to platinum (NCCH-1811). Future Sci OA. 7:FSO7212021. View Article : Google Scholar : PubMed/NCBI | |
|
Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, Wang L, Li P, Zhao Y, Duan W, et al: Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 18:432019. View Article : Google Scholar : PubMed/NCBI | |
|
Buyana B, Naki T, Alven S and Aderibigbe BA: Nanoparticles loaded with platinum drugs for colorectal cancer therapy. Int J Mol Sci. 23:112612022. View Article : Google Scholar : PubMed/NCBI | |
|
Hicks LD, Hyatt JL, Stoddard S, Tsurkan L, Edwards CC, Wadkins RM and Potter PM: Improved, selective, human intestinal carboxylesterase inhibitors designed to modulate 7-ethyl-10-[4-(1-piperidino)-1-piperidino]carbonyloxycamptothecin (Irinotecan; CPT-11) toxicity. J Med Chem. 52:3742–3752. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Meisenberg C, Ashour ME, El-Shafie L, Liao C, Hodgson A, Pilborough A, Khurram SA, Downs JA, Ward SE and El-Khamisy SF: Epigenetic changes in histone acetylation underpin resistance to the topoisomerase I inhibitor irinotecan. Nucleic Acids Res. 45:1159–1176. 2017.PubMed/NCBI |