Research advances of MAL family members in tumorigenesis and tumor progression (Review)
- Authors:
- Published online on: February 14, 2024 https://doi.org/10.3892/mmr.2024.13181
- Article Number: 57
-
Copyright: © Li et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
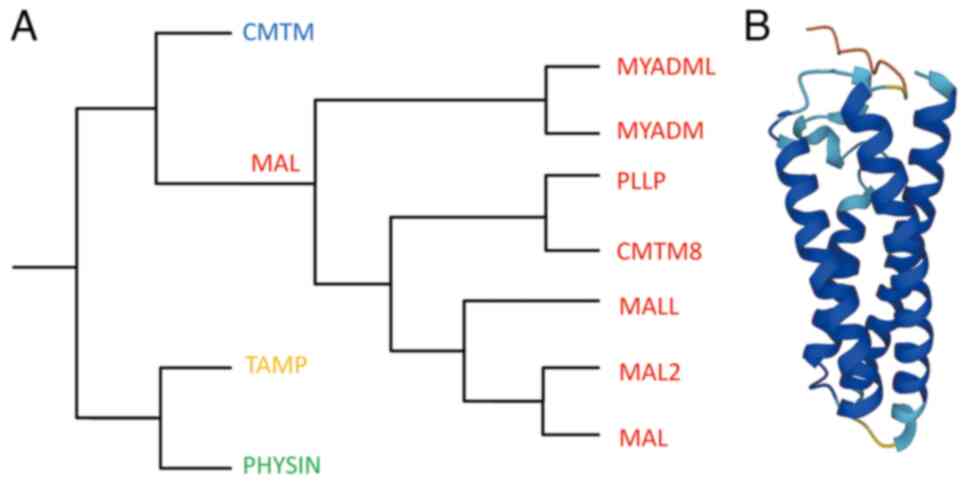
The myelin and lymphocyte protein (MAL) family belongs to the MAL and related proteins for vesicle formation and membrane link (MARVEL) superfamily, first characterized in 2002 (Fig. 1A) (1,2). Alonso and Weissman (3) first identified the human MAL cDNA while searching for genes selectively expressed during T cell differentiation. MAL has been demonstrated as an element of the machinery that transports apical proteins through direct pathways in Madin-Darby canine kidney cells depleted of endogenous MAL (4–7). At a steady state, MAL predominantly localizes to the apical zone of polarized epithelia and continuously shuttles between the Golgi apparatus and plasma membrane, functioning as a key carrier in membrane signaling in the direct transcytosis pathway (8,9).
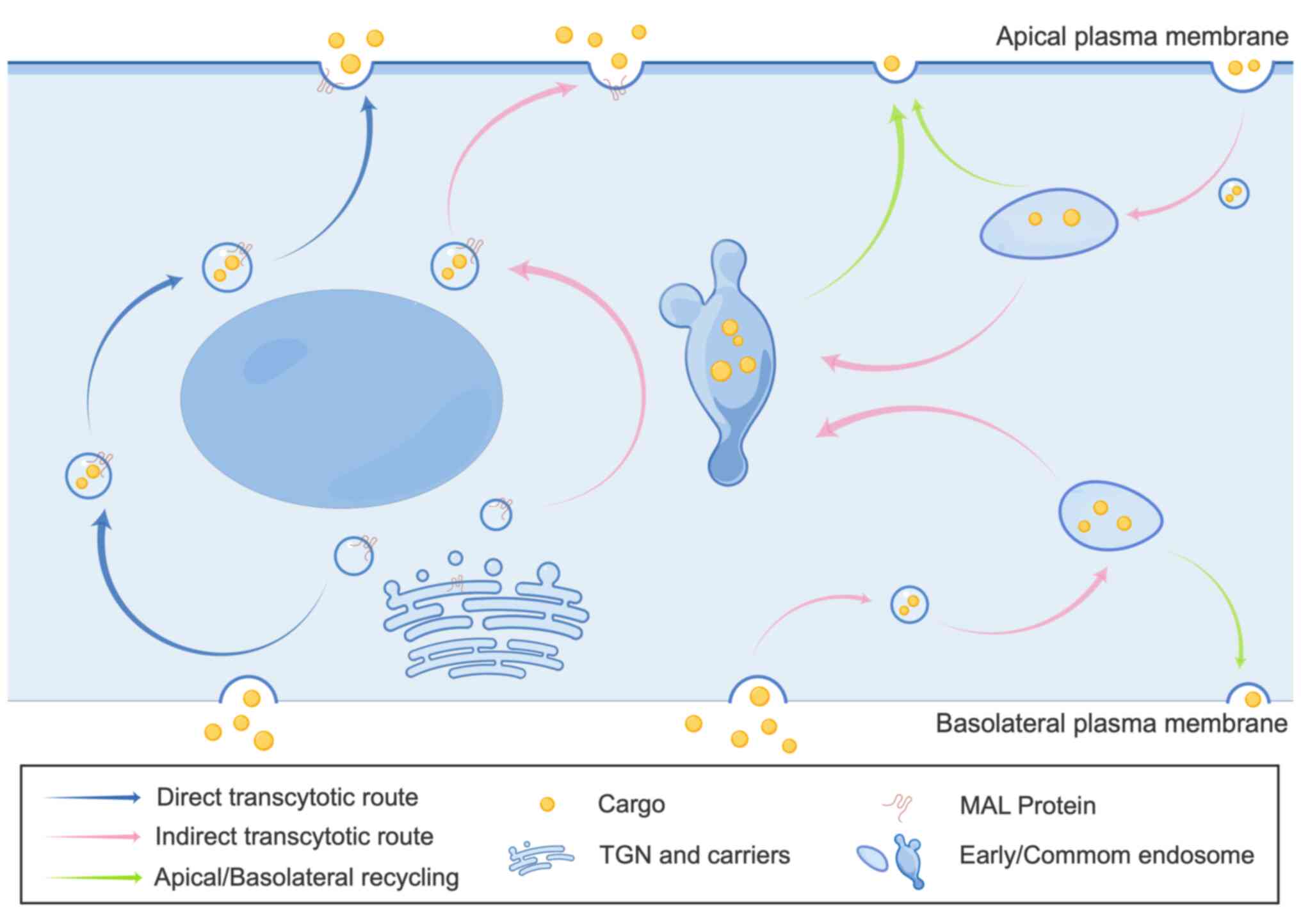
Transcytosis is a specialized transcellular transport process enabling targeted transport of cargo, such as soluble molecules, macromolecules and pathogens, across epithelial barriers via direct and indirect pathways (10). The indirect pathway is common in most polarized epithelia, while the direct pathway is exclusive to certain epithelial cell types. The direct pathway appears mediated by incorporation of cargo proteins into specialized membrane microdomains or lipid rafts that generate apical-destined vesicular carriers (11). In the direct pathway, cargo is endocytosed on one epithelial side, transported in vesicles formed through the Golgi and delivered subsequently to the opposite side of the epithelial barrier (10,11). Conversely, the indirect pathway first shuttles cargo to the basolateral or apical membrane (chiefly basolateral), where it is endocytosed into early endosomes before targeting the opposite membrane surface (11,12). Endocytosed proteins can be recycled to the original membrane, degraded in late endosomes and lysosomes, or transcytosed to the opposite surface (Fig. 2) (11,12).
The MARVEL superfamily consists of proteins containing the evolutionarily conserved MARVEL domain, present in 24 human proteins (2). Besides the MAL family, the MARVEL superfamily also comprises the chemokine-like factor MARVEL transmembrane domain containing (CMTM), physin and tight junction-associated MARVEL protein families (Fig. 1B) (1,13–17).
The MAL family is generally considered to consist of six members based on MAL proteins. MAL2, T-cell differentiation protein-like (MALL)/BENE and plasmolipin (PLLP) exhibit a tetraspanin topology similar to MAL. The other two members, myeloid differentiation-associated marker (MYADM) and MYADM-like2 (MYADML2), contain an additional MARVEL domain and form a distinct branch within the family (15). Additionally, the chemokine-like factory (CKLF) superfamily was originally described in 2001, and CKLF-like MARVEL transmembrane domain-containing 8 (CMTM8) shares 39.3% of amino acid homology with PLLP, thus it is often classified as a MAL family member (18,19). Although MAL family members have different nomenclatures, they are considered proteolipid proteins due to the 20–40% overall amino acid identity and similar hydrophobicity profiles (Table I) (20).
Functionally, the MAL family plays an important biological role in membrane transport, impacting neurological, digestive, respiratory, genitourinary and other physiological systems through signaling pathways including Notch, ERK/MAPK and EGFR (21–27). Moreover, the MAL family is involved in the pathogenesis, progression and metastasis of various cancers, with each member exhibiting differential functions in malignancies to promote or inhibit disease advancement (28,29). Numerous digestive system and female reproductive system cancers have been linked to MAL family members through various mechanisms (30–34).
DNA methylation occurs at cytosine residues in the cytosine-guanine sequence (CpG). CpG islands, often located around the gene promoters, are genomic regions with high CpG density and G+C content >50% (35,36). Abnormal hypermethylation of CpG islands upstream of tumor suppressor genes is a main mechanism of gene inactivation in human tumorigenesis, playing an important role in pathogenesis (37). Except for MYADM and MYADML2, all MAL family genes contain a CpG island at their promoter region, making them susceptible to epigenetic regulation by DNA methylation (38).
As carriers in transcellular trafficking pathways, MAL proteins can function as either tumor suppressors or promoters, influencing cancer development (39). Methylated MAL combined with cell adhesion molecule 1 (CADM1) has been widely utilized as an early diagnostic biomarker for cervical cancer (40,41). Conversely, MAL2 participates in indirect apical transport and may modulate antitumor immunity when expressed at aberrant high or low levels in different tissues (42,43). Moreover, the homologs PLLP is expressed not only in the nervous system and kidney, but also in a number of other tissues such as thymus, testis, lung, and thyroid gland (44). MALL induces nuclear aberrations that can promote carcinogenesis in various tissues (45), while CMTM8 closely regulates EGFR signaling pathways and disease progression (27). MYADM and MYADML2 are associated with myeloid differentiation and endothelial inflammation, with upregulated expression in certain diseases (46).
Therefore, MAL family members have differential tumor-associated functions and mechanisms. Further elucidating their roles in tumor pathogenesis, progression and metastasis will significantly advance molecular detection and gene therapy, providing new hope to patients with cancer. The present review discusses the relationships between individual MAL proteins and cancers, aiming to elucidate patterns and offer alternative therapeutic strategies.
MAL/VIP17
MAL is encoded on chromosome 2q11 and produces a 17 kDa integral membrane protein found in lipid rafts (47–49). Lipid rafts are sphingolipid- and cholesterol-enriched microdomains important for segregating cell surface components and influencing membrane dynamics, trafficking, adhesion, signaling and apoptosis (50,51). In epithelial cells, MAL predominantly localizes to the trans-Golgi network (TGN), transporting vesicles to regulate protein sorting (6). MAL knockdown decreases apical vesicle transport, leading to accumulation of apical proteins in the Golgi (6). In T cells, MAL affects differentiation by modulating sorting but does not impact membrane localization of lymphocyte-specific kinase (Lck) or T cell receptor signaling (52,53). MAL interacts with the glycosylphosphatidylinositol-anchored protein CD59 and Lck, suggesting that it can bind lipids to function as a membrane adaptor (6). MAL is also a key factor in exosome secretion from human T cells (54).
Beyond T cells, MAL is expressed in polarized epithelia and myelinating cells including the kidney, stomach, colon and oligodendrocytes (55–59). In these cells, apical protein/lipid transport is essential for epithelial function, with loss of polarity associated with transformation (49). During oligodendrocytes maturation and Schwann cell myelination, MAL expression and lipid raft binding mediate polarization (59).
MAL is widely expressed in respiratory, neurological, genitourinary, gastrointestinal and endocrine/exocrine epithelial tissues (Table II) (24,60,61). It has dichotomous roles in carcinogenesis as either a tumor suppressor or progression factor (39). Evidence supporting its tumor suppressor activity includes ectopic MAL expression inhibiting growth and reducing viability of cancer cells in nude mouse models, blocking G1/S transition and increasing Fas-mediated apoptosis (32,62–67). However, MAL acts as an oncogenic factor in endometrial carcinoma and certain lymphomas, such as thymic large B cell lymphomas but not in nodal diffuse large B cell lymphomas (39,68–72).
Association between aberrant MAL gene methylation/protein expression and pathological features or clinical outcomes have been reported in multiple cancers such as colon, esophagus, breast and so on (73–75). Rescue expression experiments using 5-aza-2′-deoxycytidine (decitabine, DAC) with or without trichostatin A to inhibit DNA methylation and deacetylation have provided further evidence that hypermethylation is the predominant mechanism silencing MAL in particular malignancies (Table III) (32,41,62,63,73–115).
Several studies have demonstrated that aberrant MAL promoter methylation is associated with MAL silencing in breast, esophageal and colorectal carcinomas, and shows promise as an early biomarker alongside other markers (73–75). MAL hypermethylation is common in Barrett's-associated esophageal adenocarcinoma but not squamous cell carcinoma, suggesting utility as an adenocarcinoma-specific biomarker linked to high-risk features (74). Besides suppressing motility, aggressiveness, tumorigenicity and inducing apoptosis, MAL exerts tumor suppressive effects in esophageal cancer (32). Univariate and multivariate analyses have associated MAL methylation with poorer disease-free survival in patients with gastric cancer, highlighting its potential as an independent marker (76). Moreover, the MAL protein can inhibit gastric cancer invasion and metastasis by interfering with STAT3 phosphorylation (77). Another study detected MAL hypermethylation in 80% (49/61) of colorectal cancers and 71% (45/63) of adenomas vs. only 4% (1/23) of normal mucosa samples using methylation-specific polymerase chain reaction, indicating MAL is downregulated early during progression (73).
In breast cancer cells and 69% of primary tumors, bisulfite sequencing revealed MAL promoter CpG island hypermethylation relative to normal breast epithelia (75,78). Restoring MAL expression reduces tumor cell migration and alters lipid raft organization (75,78). Among patients with breast cancer not receiving chemotherapy, low MAL expression is prognostic for worse disease-free survival, supporting its use as an adjuvant predictor (75).
CADM1/MAL methylation in high-risk human papillomavirus (HPV) positive Pap smears associates with extent and duration of underlying cervical pathology, increasing in invasive cervical cancer (79,80). Combined detection plays an important diagnostic role in identifying precancerous cervical intraepithelial neoplasia (81–83). MAL also serves as an indicator distinguishing long-term and short-term ovarian cancer prognoses, with silencing conferring treatment resistance (84,85). Lee et al (86) found that MAL methylation status marks platinum sensitivity in ovarian cancer, suggesting MAL represents a therapeutic target. Furthermore, low CADM1/high MAL levels associate with improved prognosis in Merkel cell carcinoma (87), whereas low MAL predicts poor Wilms' tumor outlook by altering the microenvironment (88). Finally, MAL demonstrates consistently reduced expression in head/neck and oral squamous cell carcinomas, with overexpression inhibiting proliferation, invasion and tumorigenesis (63,89–91,115).
Non-coding mRNAs (ncRNAs) modulate the expression of protein-coding genes and are classified as short, such as micro RNAs (miRNAs), or long ncRNA (lncRNA). LncRNA-AC103563.8 promotes oral carcinoma development by suppressing MAL expression or interacting with other tumor-related proteins, such as RPS3A, hnRNPK, HSPA9, RPS3, NCL and RPL12 (105).
MAL2
MAL2, encoded on chromosome 8q24, was first identified by Wilson et al (116) in 2001. MAL2 protein is a 19 kDa, four-transmembrane integral protein sharing 35.8% homology with the MAL proteolipid required for apical transport, considered to be the closest family member to the MAL protein (116). In hepatoma HepG2, MAL2 selectively localizes to cholesterol-rich lipid raft membrane microdomains and is crucial for indirect route of raft-dependent apical membrane transport (42). MAL2 interacts with the constitutively active, Golgi-associated serine/threonine kinase 16 to sort soluble secretory cargo through the constitutive secretory pathway at the TGN in polarized hepatocytes (117). Unlike the Golgi-predominant distribution of MAL, immunohistochemistry (IHC) of thyroid follicles indicated that MAL2 localizes to the apical membrane within lipid rafts, implicating MAL2 in transcytotic cargo transport from perinuclear endosomes to the apical surface via a raft-dependent pathway (118). However, studies in PC-3 prostate and breast cancer cells demonstrated additional MAL2 distribution in non-lipid raft components, suggesting its distinct functions inside and outside the lipid raft (119,120). In HepG2 cells, after CD59 endocytosis, some MAL2 redistributed into vesicular clusters concentrating CD59 and leaving CD59 accessible to the basolateral recycling transferrin receptor. The receptor then segregates before the clusters fuse into MAL2+ structures that move apically to deliver CD59 (21). MAL2 loss blocks apical transport of polymeric immunoglobulin A receptor (pIgA-R) and CD59, leading to perinuclear endosomal accumulation reachable by transferrin (42). MAL2 also regulates pIgA-R Golgi-to-membrane transfer, whereby pIgA-R remains in the Golgi when expressed alone in hepatic WIF-B cells, but reaches the cell surface, undergoes endocytosis and localizes to MAL2+ regions when co-expressed with MAL2 (121).
IHC shows MAL2 expression in the respiratory, digestive, genitourinary, endocrine and exocrine epithelia (often apically) as well as in specialized secretory cell clusters such as pancreatic endocrine cells. Peripheral neurons, mast cells, dendritic cells and hepatocytes also express MAL2 (Table II) (22). Chromosome 8q24 gains, encompassing the MAL2 locus, associate with several epithelial cancer types and may explain upregulated MAL2 transcription in subsets of these malignancies (122,123).
Overexpression of MAL2 in breast cancer can interact with β-catenin in breast cells, inducing c-Myc to promote proliferation and invasion by regulating epithelial-mesenchymal transition (124,125). MAL2 also associates with decreased immune infiltration and eosinophil/dendritic cell expression, conferring worse prognosis (126). Multi-omics analysis and cytology experiments by Yuan et al (127) showed high MAL2 levels in invasive breast, pancreatic, bladder, ovarian, cervical and other carcinomas, and high MAL2 expression associated with unfavorable prognosis in certain tumors. Yeast two-hybridization, pull-down and coimmunoprecipitation experiments showed that the N-terminus of MAL2 interacts with tumor protein D52 (TPD52) in breast cancer cells per (116). Jeong et al (128) demonstrated that MAL2 plays key roles in breast cancer lipid raft formation, HER2 signaling and membrane stability. As a chaperone protein for tumor-associated protein mucin 1 (MUC1), MAL2 binds MUC1 in non-raft fractions, potentially promoting breast tumorigenesis by modulating MUC1 expression and localization (120). In addition, MAL2 can prompt breast cancer immune evasion through MHC-I endocytosis and degradation, hindering antigen presentation and CD8+ T cell response. MAL2 inhibition conversely enhances cytotoxicity and recognition to suppress tumor growth (43,129). Zhu et al (130) found that the MAL2/MUC1-C/PI3K/AKT/mTOR signaling elicits triple-negative breast cancer (TNBC) aggressiveness, mitigated by targeted small molecules.
In non-small cell lung cancer cells, MAL2 overexpression hyperactivates MAPK/mTOR signaling, thus, targeting this pathway may improve therapeutic efficacy in these patients (29,131). MAL2 also interacts with IQGAP1 in pancreatic cancer, heightening ERK1/2 phosphorylation to drive progression and associate with increased metastasis (25,131,132). Chronic pancreatitis maintains MAL2 expression, providing utility as a diagnostic marker (133). The ST8SIA6-AS1/miR-145-5P/MAL2 axis promotes the progression of cholangiocarcinoma and may help improve clinical outcomes (134). MAL2 is expressed in prostate cancer and may regulate disease progression through the Notch signaling pathway (26). Gao et al (134) found that miR-129 negatively regulates expression of MAL2 in papillary thyroid carcinoma and may be a potential therapeutic target. The high expression of MAL2 exhibits TPD52-associated expression in ovarian/colorectal tumors, although the survival of patients with ovarian cancer shows no clear association (30,135,136). Various other malignancies overexpress MAL2, including gastric, cervical, bladder, oral and head/neck squamous cell carcinoma (31,34,137–139).
Circular (circ) RNAs, which are a subclass of lncRNA structured in a loop with the 3′ and 5′ RNA ends joined covalently, can inhibit miRNA activity, whereas other lncRNAs promote miRNA functions (140,141). For instance, miR-129 suppresses MAL2 in papillary thyroid cancer, representing a potential therapeutic target (140). The ST8SIA6-AS1/miR-145-5P/MAL2 axis promotes the progression of cholangiocarcinoma and may help improve clinical outcomes (142). Additional examples include co-regulation of MAL2 by miR-802 and the circRNA, circ_0084904 in cervical cancer (31), by miRNA320a alongside the lncRNA, metastasis-associated lung adenocarcinoma transcript 1 in bladder cancer (34) and by LINC00460 and miRNA320a to enable breast cancer cell proliferation and ferroptosis evasion (143).
Notably, the research of López-Coral et al (122) showed lower MAL2 protein levels in hepatocellular carcinoma, cholangiocarcinoma and renal cell carcinoma relative to normal tissue. This implies MAL2 protein may have tumor-suppressive roles, potentially by inducing actin-remodeling filopodia to reduce migration, invasion and proliferation-effects reversed upon MAL2 loss (Table IV) (122).
PLLP/TM4SF11
PLLP protein is a transmembrane protein encoded by the PLLP gene, also known as transmembrane 4 superfamily member 11 (TM4SF11), which plays a role in epithelial development, differentiation and migration (144–147). PLLP is chemically similar to MAL and proteolipid protein (PLP), a type of myelin protein, sharing 29% homology and 49% similarity with MAL. This conserved sequence facilitates the classification PLLP as a MAL family member (2,148,149). In polarized cells, PLLP predominantly localizes to the apical membrane with some basolateral distribution. PLLP, like its homologue MAL, is isolated in the lipid raft in the trans-Golgi apparatus network and before its transport to the apical and basolateral cell surfaces (147,150–152). PLLP is delivered to the plasma membrane via microtubules as a component of vesicles (147,150). Endocytosed PLLP then forms marginal vesicles transported back to the Golgi and other regions, completing an intracellular cycle (144,153,154). Interaction with ganglioside GM1 restructures the extracellular loops of PLLP, propagating a conformational signal through the plasma membrane to the intracellular domain, consistent with the role of PLLP in signal transduction (155).
Western blotting experiments demonstrate PLLP expression in nervous, digestive (stomach, esophagus and colon), renal, cardiac, pulmonary, musculoskeletal, immune (thymus) and reproductive (ovarian and testicular) tissues, as well as endocrine glands such as the adrenal, parotid, submandibular, Cowper's and prostate (44,156–158). Abundant apical localization manifests in kidney tubular epithelia and diverse gastric glandular regions (44,158). Nervous system expression includes spinal leucoplast, peripheral Schwann cells and central oligodendrocytes (44,156,157,159). While PLLP resides apically in epithelia, phosphorylated PLLP in neural cells contributes to myelination by inducing myelin precursor domains in the Golgi (44,158). PLLP plays an important role by activating the Notch signaling pathway, which is essential for cell differentiation and processes such as epidermal regeneration and diabetic wound healing (145,160). Reduced PLLP levels in patients with idiopathic pulmonary fibrosis implies protective roles in promoting endothelial development, membranes and cell junctions (161). However, no clear evidence elucidates PLLP-mediated tumorigenesis via Notch or other pathways.
MALL/BENE
The MALL protein, also called BENE, was originally identified proximal to immunoglobulin light chain κ locus. MALL comprises a protein-lipid with a four-transmembrane topology resembling PLP, PLLP and MAL that circulates between cell membranes, endosomes and Golgi to mediate apical transport (162,163). Electron microscopy and immunofluorescence analyses in endothelial-like ECV304 cells revealed predominant MALL localization to intracellular tubulovesicular structures with partial caveolin-1 colocalization (163). Co-immunoprecipitations confirmed MALL-caveolin-1 interactions, reflecting roles in cholesterol regulation (163). Beyond this membrane form, MALL also resides within promyelocytic leukemia nuclear body condensates (164). During mitosis, MALL accumulates in solid-like condensates around the spindle but, when in excess, the condensates mis-localized, altered the distribution of nuclear proteins emerin LAP2β and BAF, and caused nuclear aberrations, which are a hallmark of cancer cells (164).
MALL is expressed in prostate, intestinal, cardiac and other tissues, but not brain, thymus, hepatic or splenic tissue (15). Oncogenesis appears to induce MALL expression in some cancers such as pancreatic and kidney while it is reduced in other malignancies such as colorectal, breast and lung (164). In colorectal cancer, significantly decreased MALL impacts caveolin-1 signal transduction and Akt-1 activity (165–167). MALL suppresses colorectal growth and metastasis by inhibiting the metastasis/angiogenesis-associated ERK/MAPK pathway (167). Conversely, in pancreatic cancer there is overexpression of MALL and nuclear abnormalities conferring poorer prognosis (164). MALL also associates with unfavorable kidney cancer outcomes (168).
MYADM and MYADML2
MYADM is an eight-transmembrane protein with two MARVEL domains residing in nuclear and cytoplasmic membranes. It regulates plasma membrane-cytoskeleton connections, thereby controlling endothelial inflammation (46,159,169). Previous studies demonstrate selective MYADM expression in myeloid lineage and hematopoietic cells, implicating roles in myeloid differentiation (170–172). MYADM also shows abundant expression in various tissue epithelia and neural/pulmonary tissues (46,173). In pulmonary arterial hypertension, MYADM elicits smooth muscle proliferation through microRNA-182-3p induction and vascular remodeling (174).
Several cancers exhibit upregulated MYADM protein, including metastatic melanoma and hepatocellular carcinoma, where it constitutes an independent survival/prognostic factor (175–178). During prostate cancer metastasis, MYADM upregulation in tumor-osteoclast/endothelial co-cultures also support roles in facilitating spread (179). Furthermore, increased MYADM mRNA marks myeloid leukemia differentiation, providing utility as a disease monitoring marker (170). In biomarker studies of prostate cancer recurring within five years in African Americans, MYADM associates with this aggressive disease course (180).
MYADML2, a protein structurally similar to MYADM, exhibits elevated mRNA expression levels in hepatocellular carcinoma (181). Pathological results show cell population formed highly metastatic tumors in lung after being mutagenized with CRISPR activation. In vitro validation indicated overexpression of MYADML2 promoted proliferation and invasion of cells, and the inhibition suppressed cancer progress (181). Its role in reducing sensitivity to chemotherapeutic drugs has also been documented (181).
CMTM8
CMTM8, a novel chemokine comprising 173 amino acids, shares up to 39.3% amino acid sequence homology with the MAL family molecule PLLP, thus classifying it within the MAL family (18). Research has demonstrated that CMTM8 expedites the internalization of transferrin receptor and EGFR, hastening the clearance of EGFR from the cell surface upon ligand induction (182). Furthermore, CMTM8 modulates EGFR-mediated signaling pathways by reducing ERK phosphorylation levels (183). Subsequent investigations have revealed that CMTM8 induces apoptosis in cells through both caspase-dependent and caspase-independent pathways (184). Li et al (182) reported CMTM8-V2 as a selective splicing isoform of CMTM8, maintaining the ability to induce apoptosis. However, the second exon encoding the MARVEL domain and cytoplasmic YXX motif is absent in this isoform, thereby not impacting EGFR internalization.
While CMTM8 is widely expressed in numerous healthy human tissues, its downregulation or deletion has been observed in several solid tumors, including gastric cancer, lung squamous cell carcinoma, cervical cancer and renal clear cell carcinoma (182,185–187). Overexpression of CMTM8 induces apoptosis in hepatocellular carcinoma cells by influencing caspase expression through mitochondria, thereby mediating related signaling pathways (184). Conversely, downregulation of CMTM8 activates the c-Met signaling pathway, leading to the epithelial-mesenchymal transition of hepatocellular carcinoma cells, facilitating tumor migration and invasion (188). In TNBC, miR-582-5p selectively inhibits CMTM8, resulting in decreased CMTM8 expression and attenuating its inhibitory effect on TNBC cell migration and invasion (189). Moreover, CMTM8 has been shown to inhibit the EGFR signaling pathway activity in osteosarcoma, suggesting its potential role as an osteosarcoma suppressor gene (190). Additionally, upregulation of CMTM8 inhibits the proliferation and invasion of bladder cancer T24 cells and enhances their sensitivity to chemotherapeutic drugs (191).
However, Gao et al (192) reported that overexpression of CMTM8 in bladder cancer promoted tumor growth and metastasis. High expression of CMTM8 has also been confirmed in colon and ovarian cancer (193,194). Lastly, Shi et al (195) identified CMTM8 as a critical mediator of lysophosphatidic acid (LPA)-induced pancreatic cancer invasion. CMTM8 interacts with LPA1 to activate the carcinogenic β-catenin signaling transduction, thereby enhancing tumor migration and invasion.
Conclusion
The MAL family encompasses members that are widely distributed across various bodily systems, with their involvement in the digestive, respiratory, urinary and circulatory systems being successively uncovered. These proteins play crucial roles in tumorigenesis signaling pathways and cell cycle regulation, thereby influencing tumor development and patient prognosis (38,39,42,68,136,164). However, the current understanding of their specific molecular mechanisms remains limited. Notably, high expression of MAL family members in normal tissues and their decreased expression in tumor tissues provide a reliable basis for tumor diagnosis. Furthermore, the expression of MAL in certain tumor tissues associates with the malignancy of tumors, offering prognostic value for patients with cancer (38,85,101,127,155,164,184). Overexpression of MAL has been shown to induce apoptosis in tumor cells, while its inhibition leads to epithelial-to-mesenchymal transition, suggesting its potential as a suppressor of tumor cell proliferation and a candidate for tumor immunotherapy (1,38).
Non-invasive molecular detection techniques for cancer screening are rapidly advancing, with MAL family members serving as important cancer biomarkers in clinical testing (Table V). These members have been extensively utilized in non-invasive tests, such as MAL methylation assays in blood, urine and feces-derived samples from patients with various tumors (82,196–199), as well as the analysis of MAL2 transcript levels in blood from patients with gynecological and metastatic breast cancer (200,201). Integration of antibodies targeting MAL family members into existing antibody panels used in standard clinical practice for identifying cancer biomarkers in biopsies and surgical specimens, along with their application in liquid biopsies, may offer valuable avenues for enhanced prognostic and diagnostic insights in cancer cases.
While gene methylation or expression analyses are not commonly performed in medical pathology departments, IHC analysis of protein expression stands as the gold standard for characterizing cancer cells and has been used in research. Additionally, RT-qPCR and western blotting techniques have been employed to investigate the expression of MAL family genes.
Moreover, MAL family members can serve as predictive biomarkers for specific treatment modalities. For instance, MAL has been identified as one of the most highly expressed genes in survivors of short-term serous ovarian cancer (84), with its transcripts significantly overexpressed in ovarian cancer cell lines resistant to conventional platinum-based and other chemotherapeutic agents (85). This suggests its potential use in predicting the response to platinum-based drugs and as a target for developing novel therapies to improve the sensitivity of ovarian cancer to these drugs. Similarly, MAL serves as a potential biomarker with clinical significance in predicting the response of patients with breast cancer to anthracycline and taxane, commonly used in adjuvant chemotherapy for early breast cancer (38,202,203). Additionally, the level of MAL2 transcripts in pancreatic cancer demonstrates an inverse correlation with resistance to various chemotherapeutic agents, making it a potential indicator of chemotherapy response (204). Furthermore, MYADM has been identified as a potential biomarker for predicting the response to the drug MS-275, and possibly other histone deacetylase inhibitors, in colon adenocarcinoma (205).
While progress has been made in understanding the function of certain MAL family proteins, further exploration of their expression, molecular mechanisms and related signaling pathways in tumors is expected to not only serve as molecular markers for detecting tumorigenesis, progression and metastasis, but also lead to new breakthroughs in prognostic prediction and treatment of patients with cancer. This necessitates continued research to fully comprehend the role of MAL family proteins in normal and tumor cells, with potential implications for the development of targeted therapeutic agents.
In conclusion, a deeper investigation into the expression, molecular mechanisms and associated signaling pathways of MAL family members in tumors is anticipated to serve not only as molecular markers for detecting tumorigenesis, progression and metastasis, but also to yield novel insights for prognostic prediction and treatment of patients with cancer.
Acknowledgements
Not applicable.
Funding
The present review was supported by The National Natural Science Foundation of China (grant no. 82260555), Medical Innovation and Development Project of Lanzhou University (grant no. lzuyxcx-2022-177) and Major Science and Technology Projects of Gansu Province (grant no. 22ZD6FA021-4).
Availability of data and materials
Not applicable.
Authors' contributions
ML, YD and XZ substantially contributed to the conception and the design of the study, or in the acquisition, analysis and interpretation of the data; and ML, YD, XZ and WZ contributed to manuscript drafting or critical revisions on the intellectual content. All authors read and approved the final version of the manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Rubio-Ramos A, Labat-de-Hoz L, Correas I and Alonso MA: The MAL protein, an integral component of specialized membranes, in normal cells and cancer. Cells. 10:10652021. View Article : Google Scholar : PubMed/NCBI | |
|
Sánchez-Pulido L, Martín-Belmonte F, Valencia A and Alonso MA: MARVEL: A conserved domain involved in membrane apposition events. Trends Biochem Sci. 27:599–601. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Alonso MA and Weissman SM: cDNA cloning and sequence of MAL, a hydrophobic protein associated with human T-cell differentiation. Proc Natl Acad Sci USA. 84:1997–2001. 1987. View Article : Google Scholar : PubMed/NCBI | |
|
Cheong KH, Zacchetti D, Schneeberger EE and Simons K: VIP17/MAL, a lipid raft-associated protein, is involved in apical transport in MDCK cells. Proc Natl Acad Sci USA. 96:6241–6248. 1999. View Article : Google Scholar : PubMed/NCBI | |
|
Martín-Belmonte F, Puertollano R, Millán J and Alonso MA: The MAL proteolipid is necessary for the overall apical delivery of membrane proteins in the polarized epithelial Madin-Darby canine kidney and fischer rat thyroid cell lines. Mol Biol Cell. 11:2033–2045. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Martín-Belmonte F, Arvan P and Alonso MA: MAL mediates apical transport of secretory proteins in polarized epithelial Madin-Darby canine kidney cells. J Biol Chem. 276:49337–49342. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Puertollano R, Martín-Belmonte F, Millán J, de Marco MC, Albar JP, Kremer L and Alonso MA: The MAL proteolipid is necessary for normal apical transport and accurate sorting of the influenza virus hemagglutinin in Madin-Darby canine kidney cells. J Cell Biol. 145:141–151. 1999. View Article : Google Scholar : PubMed/NCBI | |
|
Martín-Belmonte F, Kremer L, Albar JP, Marazuela M and Alonso MA: Expression of the MAL gene in the thyroid: The MAL proteolipid, a component of glycolipid-enriched membranes, is apically distributed in thyroid follicles. Endocrinology. 139:2077–2084. 1998. View Article : Google Scholar : PubMed/NCBI | |
|
Puertollano R and Alonso MA: MAL, an integral element of the apical sorting machinery, is an itinerant protein that cycles between the trans-Golgi network and the plasma membrane. Mol Biol Cell. 10:3435–3447. 1999. View Article : Google Scholar : PubMed/NCBI | |
|
Mostov KE, Verges M and Altschuler Y: Membrane traffic in polarized epithelial cells. Curr Opin Cell Biol. 12:483–490. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Simons K and Wandinger-Ness A: Polarized sorting in epithelia. Cell. 62:207–210. 1990. View Article : Google Scholar : PubMed/NCBI | |
|
Matter K and Mellman I: Mechanisms of cell polarity: Sorting and transport in epithelial cells. Curr Opin Cell Biol. 6:545–554. 1994. View Article : Google Scholar : PubMed/NCBI | |
|
Raleigh DR, Marchiando AM, Zhang Y, Shen L, Sasaki H, Wang Y, Long M and Turner JR: Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol Biol Cell. 21:1200–1213. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Duan HJ, Li XY, Liu C and Deng XL: Chemokine-like factor-like MARVEL transmembrane domain-containing family in autoimmune diseases. Chin Med J (Engl). 133:951–958. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Magyar JP, Ebensperger C, Schaeren-Wiemers N and Suter U: Myelin and lymphocyte protein (MAL/MVP17/VIP17) and plasmolipin are members of an extended gene family. Gene. 189:269–275. 1997. View Article : Google Scholar : PubMed/NCBI | |
|
Janz R, Südhof TC, Hammer RE, Unni V, Siegelbaum SA and Bolshakov VY: Essential roles in synaptic plasticity for synaptogyrin I and synaptophysin I. Neuron. 24:687–700. 1999. View Article : Google Scholar : PubMed/NCBI | |
|
Adams DJ, Arthur CP and Stowell MH: Architecture of the synaptophysin/synaptobrevin complex: Structural evidence for an entropic clustering function at the synapse. Sci Rep. 5:136592015. View Article : Google Scholar : PubMed/NCBI | |
|
Han W, Ding P, Xu M, Wang L, Rui M, Shi S, Liu Y, Zheng Y, Chen Y, Yang T and Ma D: Identification of eight genes encoding chemokine-like factor superfamily members 1–8 (CKLFSF1-8) by in silico cloning and experimental validation. Genomics. 81:609–617. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Lu J, Wu QQ, Zhou YB, Zhang KH, Pang BX, Li L, Sun N, Wang HS, Zhang S, Li WJ, et al: Cancer research advance in CKLF-like MARVEL transmembrane domain containing member family (review). Asian Pac J Cancer Prev. 17:2741–2744. 2016.PubMed/NCBI | |
|
Pérez P, Puertollano R and Alonso MA: Structural and biochemical similarities reveal a family of proteins related to the MAL proteolipid, a component of detergent-insoluble membrane microdomains. Biochem Biophys Res Commun. 232:618–621. 1997. View Article : Google Scholar : PubMed/NCBI | |
|
de Marco MC, Puertollano R, Martínez-Menárguez JA and Alonso MA: Dynamics of MAL2 during glycosylphosphatidylinositol-anchored protein transcytotic transport to the apical surface of hepatoma HepG2 cells. Traffic. 7:61–73. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Marazuela M, Acevedo A, García-López MA, Adrados M, de Marco MC and Alonso MA: Expression of MAL2, an integral protein component of the machinery for basolateral-to-apical transcytosis, in human epithelia. J Histochem Cytochem. 52:243–252. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Muppa P, Dao L, McCann B, Mcphail E, Shi M and Rech K: Co-expression of MAL and PD-L2 by Immunohistochemistry is Specific for Primary Mediastinal Large B-Cell Lymphoma. Lab Invest. 99 (Suppl 1):972019. | |
|
Marazuela M, Acevedo A, Adrados M, García-López MA and Alonso MA: Expression of MAL, an integral protein component of the machinery for raft-mediated pical transport, in human epithelia. J Histochem Cytochem. 51:665–674. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang B, Xiao J, Cheng X and Liu T: MAL2 interacts with IQGAP1 to promote pancreatic cancer progression by increasing ERK1/2 phosphorylation. Biochem Biophys Res Commun. 554:63–70. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zheng C, Wang J, Zhang J, Hou S, Zheng Y and Wang Q: Myelin and lymphocyte protein 2 regulates cell proliferation and metastasis through the Notch pathway in prostate adenocarcinoma. Transl Androl Urol. 10:2067–2077. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhu J, Shimizu E, Zhang X, Partridge NC and Qin L: EGFR signaling suppresses osteoblast differentiation and inhibits expression of master osteoblastic transcription factors Runx2 and Osterix. J Cell Biochem. 112:1749–1760. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Bello-Morales R, Pérez-Hernández M, Rejas MT, Matesanz F, Alcina A and López-Guerrero JA: Interaction of PLP with GFP-MAL2 in the human oligodendroglial cell line HOG. PLoS One. 6:e193882011. View Article : Google Scholar : PubMed/NCBI | |
|
Lian Z, Yan X, Diao Y, Cui D and Liu H: T cell differentiation protein 2 facilitates cell proliferation by enhancing mTOR-mediated ribosome biogenesis in non-small cell lung cancer. Discov Oncol. 13:262022. View Article : Google Scholar : PubMed/NCBI | |
|
Byrne JA, Maleki S, Hardy JR, Gloss BS, Murali R, Scurry JP, Fanayan S, Emmanuel C, Hacker NF, Sutherland RL, et al: MAL2 and tumor protein D52 (TPD52) are frequently overexpressed in ovarian carcinoma, but differentially associated with histological subtype and patient outcome. BMC Cancer. 10:4972010. View Article : Google Scholar : PubMed/NCBI | |
|
Chen L, Li H, Yao D, Zou Q, Yu W and Zhou L: The novel circ_0084904/miR-802/MAL2 axis promotes the development of cervical cancer. Reprod Biol. 22:1006002022. View Article : Google Scholar : PubMed/NCBI | |
|
Mimori K, Shiraishi T, Mashino K, Sonoda H, Yamashita K, Yoshinaga K, Masuda T, Utsunomiya T, Alonso MA, Inoue H and Mori M: MAL gene expression in esophageal cancer suppresses motility, invasion and tumorigenicity and enhances apoptosis through the Fas pathway. Oncogene. 22:3463–3471. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Zorzan E, Elgendy R, Guerra G, Da Ros S, Gelain ME, Bonsembiante F, Garaffo G, Vitale N, Piva R, Marconato L, et al: Hypermethylation-mediated silencing of CIDEA, MAL and PCDH17 tumour suppressor genes in canine DLBCL: From multi-omics analyses to mechanistic studies. Int J Mol Sci. 23:40212022. View Article : Google Scholar : PubMed/NCBI | |
|
Tao L, Mu X, Chen H, Jin D, Zhang R, Zhao Y, Fan J, Cao M and Zhou Z: FTO modifies the m6A level of MALAT and promotes bladder cancer progression. Clin Transl Med. 11:e3102021. View Article : Google Scholar : PubMed/NCBI | |
|
Edwards JR, Yarychkivska O, Boulard M and Bestor TH: DNA methylation and DNA methyltransferases. Epigenetics Chromatin. 10:232017. View Article : Google Scholar : PubMed/NCBI | |
|
Deaton AM and Bird A: CpG islands and the regulation of transcription. Genes Dev. 25:1010–1022. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Wang JS, Guo M, Montgomery EA, Thompson RE, Cosby H, Hicks L, Wang S, Herman JG and Canto MI: DNA promoter hypermethylation of p16 and APC predicts neoplastic progression in Barrett's esophagus. Am J Gastroenterol. 104:2153–2160. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Labat-de-Hoz L, Rubio-Ramos A, Correas I and Alonso MA: The MAL family of proteins: Normal function, expression in cancer, and potential use as cancer biomarkers. Cancers (Basel). 15:28012023. View Article : Google Scholar : PubMed/NCBI | |
|
Lara-Lemus R: On the role of myelin and lymphocyte protein (MAL) in cancer: A puzzle with two faces. J Cancer. 10:2312–2318. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Overmeer RM, Louwers JA, Meijer CJ, van Kemenade FJ, Hesselink AT, Daalmeijer NF, Wilting SM, Heideman DA, Verheijen RH, Zaal A, et al: Combined CADM1 and MAL promoter methylation analysis to detect (pre-)malignant cervical lesions in high-risk HPV-positive women. Int J Cancer. 129:2218–2225. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Holubekova V, Mersakova S, Grendar M, Snahnicanova Z, Kudela E, Kalman M, Lasabova Z, Danko J and Zubor P: The role of CADM1 and MAL promoter methylation in inflammation and cervical intraepithelial neoplasia. Genet Test Mol Biomarkers. 24:256–263. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
de Marco MC, Martín-Belmonte F, Kremer L, Albar JP, Correas I, Vaerman JP, Marazuela M, Byrne JA and Alonso MA: MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells. J Cell Biol. 159:37–44. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Fang Y, Wang L, Wan C, Sun Y, Van der Jeught K, Zhou Z, Dong T, So KM, Yu T, Li Y, et al: MAL2 drives immune evasion in breast cancer by suppressing tumor antigen presentation. J Clin Invest. 131:e1408372021. View Article : Google Scholar : PubMed/NCBI | |
|
Hamacher M, Pippirs U, Köhler A, Müller HW and Bosse F: Plasmolipin: Genomic structure, chromosomal localization, protein expression pattern, and putative association with Bardet-Biedl syndrome. Mamm Genome. 12:933–937. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Hanahan D and Weinberg RA: Hallmarks of cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Aranda JF, Reglero-Real N, Kremer L, Marcos-Ramiro B, Ruiz-Sáenz A, Calvo M, Enrich C, Correas I, Millán J and Alonso MA: MYADM regulates Rac1 targeting to ordered membranes required for cell spreading and migration. Mol Biol Cell. 22:1252–1262. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Alonso MA, Barton DE and Francke U: Assignment of the T-cell differentiation gene MAL to human chromosome 2, region cen-q13. Immunogenetics. 27:91–95. 1988. View Article : Google Scholar : PubMed/NCBI | |
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM and Haussler D: The human genome browser at UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Marazuela M and Alonso MA: Expression of MAL and MAL2, two elements of the protein machinery for raft-mediated transport, in normal and neoplastic human tissue. Histol Histopathol. 19:925–933. 2004.PubMed/NCBI | |
|
Michel V and Bakovic M: Lipid rafts in health and disease. Biol Cell. 99:129–140. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Patra SK and Bettuzzi S: Epigenetic DNA-methylation regulation of genes coding for lipid raft-associated components: A role for raft proteins in cell transformation and cancer progression (review). Oncol Rep. 17:1279–1290. 2007.PubMed/NCBI | |
|
Leitner J, Mahasongkram K, Schatzlmaier P, Pfisterer K, Leksa V, Pata S, Kasinrerk W, Stockinger H and Steinberger P: Differentiation and activation of human CD4 T cells is associated with a gradual loss of myelin and lymphocyte protein. Eur J Immunol. 51:848–863. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Antón OM, Andrés-Delgado L, Reglero-Real N, Batista A and Alonso MA: MAL protein controls protein sorting at the supramolecular activation cluster of human T lymphocytes. J Immunol. 186:6345–6356. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Ventimiglia LN, Fernández-Martín L, Martínez-Alonso E, Antón OM, Guerra M, Martínez-Menárguez JA, Andrés G and Alonso MA: Cutting edge: Regulation of exosome secretion by the integral MAL protein in T cells. J Immunol. 195:810–814. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Kim T, Fiedler K, Madison DL, Krueger WH and Pfeiffer SE: Cloning and characterization of MVP17: A developmentally regulated myelin protein in oligodendrocytes. J Neurosci Res. 42:413–422. 1995. View Article : Google Scholar : PubMed/NCBI | |
|
Schaeren-Wiemers N, Schaefer C, Valenzuela DM, Yancopoulos GD and Schwab ME: Identification of new oligodendrocyte- and myelin-specific genes by a differential screening approach. J Neurochem. 65:10–22. 1995. View Article : Google Scholar : PubMed/NCBI | |
|
Zacchetti D, Peränen J, Murata M, Fiedler K and Simons K: VIP17/MAL, a proteolipid in apical transport vesicles. FEBS Lett. 377:465–469. 1995. View Article : Google Scholar : PubMed/NCBI | |
|
Millán J, Puertollano R, Fan L and Alonso MA: Caveolin and MAL, two protein components of internal detergent-insoluble membranes, are in distinct lipid microenvironments in MDCK cells. Biochem Biophys Res Commun. 233:707–712. 1997. View Article : Google Scholar : PubMed/NCBI | |
|
Frank M, Schaeren-Wiemers N, Schneider R and Schwab ME: Developmental expression pattern of the myelin proteolipid MAL indicates different functions of MAL for immature Schwann cells and in a late step of CNS myelinogenesis. J Neurochem. 73:587–597. 1999. View Article : Google Scholar : PubMed/NCBI | |
|
Frank M: MAL, a proteolipid in glycosphingolipid enriched domains: Functional implications in myelin and beyond. Prog Neurobiol. 60:531–544. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Copie-Bergman C, Plonquet A, Alonso MA, Boulland ML, Marquet J, Divine M, Möller P, Leroy K and Gaulard P: MAL expression in lymphoid cells: further evidence for MAL as a distinct molecular marker of primary mediastinal large B-cell lymphomas. Mod Pathol. 15:1172–1180. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Overmeer RM, Henken FE, Bierkens M, Wilting SM, Timmerman I, Meijer CJ, Snijders PJ and Steenbergen RD: Repression of MAL tumour suppressor activity by promoter methylation during cervical carcinogenesis. J Pathol. 219:327–336. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Cao W, Zhang ZY, Xu Q, Sun Q, Yan M, Zhang J, Zhang P, Han ZG and Chen WT: Epigenetic silencing of MAL, a putative tumor suppressor gene, can contribute to human epithelium cell carcinoma. Mol Cancer. 9:2962010. View Article : Google Scholar : PubMed/NCBI | |
|
Wilting SM, de Wilde J, Meijer CJ, Berkhof J, Yi Y, van Wieringen WN, Braakhuis BJ, Meijer GA, Ylstra B, Snijders PJ and Steenbergen RD: Integrated genomic and transcriptional profiling identifies chromosomal loci with altered gene expression in cervical cancer. Genes Chromosomes Cancer. 47:890–905. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Maruya S, Kim HW, Weber RS, Lee JJ, Kies M, Luna MA, Batsakis JG and El-Naggar AK: Gene expression screening of salivary gland neoplasms: Molecular markers of potential histogenetic and clinical significance. J Mol Diagn. 6:180–190. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Suzuki M, Shiraishi K, Eguchi A, Ikeda K, Mori T, Yoshimoto K, Ohba Y, Yamada T, Ito T, Baba Y and Baba H: Aberrant methylation of LINE-1, SLIT2, MAL and IGFBP7 in non-small cell lung cancer. Oncol Rep. 29:1308–1314. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Ostrow KL, Park HL, Hoque MO, Kim MS, Liu J, Argani P, Westra W, Van Criekinge W and Sidransky D: Pharmacologic unmasking of epigenetically silenced genes in breast cancer. Clin Cancer Res. 15:1184–1191. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Li D, Zhang J, Wu L, Yang X, Chen Z and Yuan J: Myelin and lymphocyte protein (MAL): A novel biomarker for uterine corpus endometrial carcinoma. Cancer Manag Res. 13:7311–7323. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
His ED, Sup SJ, Alemany C, Tso E, Skacel M, Elson P, Alonso MA and Pohlman B: MAL is expressed in a subset of Hodgkin lymphoma and identifies a population of patients with poor prognosis. Am J Clin Pathol. 125:776–782. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Kohno T, Moriuchi R, Katamine S, Yamada Y, Tomonaga M and Matsuyama T: Identification of genes associated with the progression of adult T cell leukemia (ATL). Jpn J Cancer Res. 91:1103–1110. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Chan JK: Mediastinal large B-cell lymphoma: New evidence in support of its distinctive identity. Adv Anat Pathol. 7:201–209. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Copie-Bergman C, Gaulard P, Maouche-Chrétien L, Brière J, Haioun C, Alonso MA, Roméo PH and Leroy K: The MAL gene is expressed in primary mediastinal large B-cell lymphoma. Blood. 94:3567–3575. 1999. View Article : Google Scholar : PubMed/NCBI | |
|
Lind GE, Ahlquist T, Kolberg M, Berg M, Eknaes M, Alonso MA, Kallioniemi A, Meling GI, Skotheim RI, Rognum TO, et al: Hypermethylated MAL gene-a silent marker of early colon tumorigenesis. J Transl Med. 6:132008. View Article : Google Scholar : PubMed/NCBI | |
|
Jin Z, Wang L, Zhang Y, Cheng Y, Gao Y, Feng X, Dong M, Cao Z, Chen S, Yu H, et al: MAL hypermethylation is a tissue-specific event that correlates with MAL mRNA expression in esophageal carcinoma. Sci Rep. 3:28382013. View Article : Google Scholar : PubMed/NCBI | |
|
Horne HN, Lee PS, Murphy SK, Alonso MA, Olson JA Jr and Marks JR: Inactivation of the MAL gene in breast cancer is a common event that predicts benefit from adjuvant chemotherapy. Mol Cancer Res. 7:199–209. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Buffart TE, Overmeer RM, Steenbergen RD, Tijssen M, van Grieken NC, Snijders PJ, Grabsch HI, van de Velde CJ, Carvalho B and Meijer GA: MAL promoter hypermethylation as a novel prognostic marker in gastric cancer. Br J Cancer. 99:1802–1807. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Geng Z, Li J, Li S, Wang Y, Zhang L, Hu Q, Wang X, Zuo L, Song X, Zhang X, et al: MAL protein suppresses the metastasis and invasion of GC cells by interfering with the phosphorylation of STAT3. J Transl Med. 20:502022. View Article : Google Scholar : PubMed/NCBI | |
|
Deng F and Han Bae Y: Lipid raft-mediated and upregulated coordination pathways assist transport of glycocholic acid-modified nanoparticle in a human breast cancer cell line of SK-BR-3. Int J Pharm. 617:1215892022. View Article : Google Scholar : PubMed/NCBI | |
|
Bierkens M, Hesselink AT, Meijer CJ, Heideman DA, Wisman GB, van der Zee AG, Snijders PJ and Steenbergen RD: CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer. 133:1293–1299. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Salta S, Lobo J, Magalhães B, Henrique R and Jerónimo C: DNA methylation as a triage marker for colposcopy referral in HPV-based cervical cancer screening: A systematic review and meta-analysis. Clin Epigenetics. 15:1252023. View Article : Google Scholar : PubMed/NCBI | |
|
De Strooper LM, Hesselink AT, Berkhof J, Meijer CJ, Snijders PJ, Steenbergen RD and Heideman DA: Combined CADM1/MAL methylation and cytology testing for colposcopy triage of high-risk HPV-positive women. Cancer Epidemiol Biomarkers Prev. 23:1933–1937. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Leffers M, Herbst J, Kropidlowski J, Prieske K, Bohnen AL, Peine S, Jaeger A, Oliveira-Ferrer L, Goy Y, Schmalfeldt B, et al: Combined liquid biopsy methylation analysis of CADM1 and MAL in cervical cancer patients. Cancers (Basel). 14:39542022. View Article : Google Scholar : PubMed/NCBI | |
|
Phillips S, Cassells K, Garland SM, Machalek DA, Roberts JM, Templeton DJ, Jin F, Poynten IM, Hillman RJ, Grulich AE, et al: Gene methylation of CADM1 and MAL identified as a biomarker of high grade anal intraepithelial neoplasia. Sci Rep. 12:35652022. View Article : Google Scholar : PubMed/NCBI | |
|
Berchuck A, Iversen ES, Lancaster JM, Pittman J, Luo J, Lee P, Murphy S, Dressman HK, Febbo PG, West M, et al: Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Cancer Res. 11:3686–3696. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Zanotti L, Romani C, Tassone L, Todeschini P, Tassi RA, Bandiera E, Damia G, Ricci F, Ardighieri L, Calza S, et al: MAL gene overexpression as a marker of high-grade serous ovarian carcinoma stem-like cells that predicts chemoresistance and poor prognosis. BMC Cancer. 17:3662017. View Article : Google Scholar : PubMed/NCBI | |
|
Lee PS, Teaberry VS, Bland AE, Huang Z, Whitaker RS, Baba T, Fujii S, Secord AA, Berchuck A and Murphy SK: Elevated MAL expression is accompanied by promoter hypomethylation and platinum resistance in epithelial ovarian cancer. Int J Cancer. 126:1378–1389. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Iwasaki T, Matsushita M, Nonaka D, Nagata K, Kato M, Kuwamoto S, Murakami I and Hayashi K: Lower expression of CADM1 and higher expression of MAL in Merkel cell carcinomas are associated with Merkel cell polyomavirus infection and better prognosis. Hum Pathol. 48:1–8. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Su C, Huang R, Yu Z, Zheng J, Liu F, Liang H and Mo Z: Myelin and lymphocyte protein serves as a prognostic biomarker and is closely associated with the tumor microenvironment in the nephroblastoma. Cancer Med. 11:1427–1438. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Pal SK, Noguchi S, Yamamoto G, Yamada A, Isobe T, Hayashi S, Tanaka J, Tanaka Y, Kamijo R, Yamane GY and Tachikawa T: Expression of myelin and lymphocyte protein (MAL) in oral carcinogenesis. Med Mol Morphol. 45:222–228. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Beder LB, Gunduz M, Hotomi M, Fujihara K, Shimada J, Tamura S, Gunduz E, Fukushima K, Yaykasli K, Grenman R, et al: T-lymphocyte maturation-associated protein gene as a candidate metastasis suppressor for head and neck squamous cell carcinomas. Cancer Sci. 100:873–880. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Luo Y, Zhou LQ, Yang F, Chen JC, Chen JJ and Wang YJ: Construction and analysis of a conjunctive diagnostic model of HNSCC with random forest and artificial neural network. Sci Rep. 13:67362023. View Article : Google Scholar : PubMed/NCBI | |
|
Yue Y, Song M, Qiao Y, Li P, Yuan Y, Lian J, Wang S and Zhang Y: Gene function analysis and underlying mechanism of esophagus cancer based on microarray gene expression profiling. Oncotarget. 8:105222–105237. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Wang X, Li G, Luo Q and Gan C: Identification of crucial genes associated with esophageal squamous cell carcinoma by gene expression profile analysis. Oncol Lett. 15:8983–8990. 2018.PubMed/NCBI | |
|
Visser E, Franken IA, Brosens LA, Ruurda JP and van Hillegersberg R: Prognostic gene expression profiling in esophageal cancer: A systematic review. Oncotarget. 8:5566–5577. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Choi B, Han TS, Min J, Hur K, Lee SM, Lee HJ, Kim YJ and Yang HK: MAL and TMEM220 are novel DNA methylation markers in human gastric cancer. Biomarkers. 22:35–44. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Kurashige J, Sawada G, Takahashi Y, Eguchi H, Sudo T, Ikegami T, Yoshizumi T, Soejima Y, Ikeda T, Kawanaka H, et al: Suppression of MAL gene expression in gastric cancer correlates with metastasis and mortality. Fukuoka Igaku Zasshi. 104:344–349. 2013.PubMed/NCBI | |
|
Ahlquist T, Lind GE, Costa VL, Meling GI, Vatn M, Hoff GS, Rognum TO, Skotheim RI, Thiis-Evensen E and Lothe RA: Gene methylation profiles of normal mucosa, and benign and malignant colorectal tumors identify early onset markers. Mol Cancer. 7:942008. View Article : Google Scholar : PubMed/NCBI | |
|
Patai ÁV, Valcz G, Hollósi P, Kalmár A, Péterfia B, Patai Á, Wichmann B, Spisák S, Barták BK, Leiszter K, et al: Comprehensive DNA methylation analysis reveals a common ten-gene methylation signature in colorectal adenomas and carcinomas. PLoS One. 10:e01338362015. View Article : Google Scholar : PubMed/NCBI | |
|
Sambuudash O, Kim HS and Cho MY: Lack of aberrant methylation in an adjacent area of left-sided colorectal cancer. Yonsei Med J. 58:749–755. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Liu X, Bi H, Ge A, Xia T, Fu J, Liu Y, Sun H, Li D and Zhao Y: DNA hypermethylation of MAL gene may act as an independent predictor of favorable prognosis in patients with colorectal cancer. Transl Cancer Res. 8:1985–1996. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Ma R, Xu YE, Wang M and Peng W: Suppression of MAL gene expression is associated with colorectal cancer metastasis. Oncol Lett. 10:957–961. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Huang KT, Mikeska T, Li J, Takano EA, Millar EK, Graham PH, Boyle SE, Campbell IG, Speed TP, Dobrovic A and Fox SB: Assessment of DNA methylation profiling and copy number variation as indications of clonal relationship in ipsilateral and contralateral breast cancers to distinguish recurrent breast cancer from a second primary tumour. BMC Cancer. 15:6692015. View Article : Google Scholar : PubMed/NCBI | |
|
Vasiljević N, Scibior-Bentkowska D, Brentnall AR, Cuzick J and Lorincz AT: Credentialing of DNA methylation assays for human genes as diagnostic biomarkers of cervical intraepithelial neoplasia in high-risk HPV positive women. Gynecol Oncol. 132:709–714. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Ki EY, Lee KH, Hur SY, Rhee JE, Kee MK, Kang C and Park JS: Methylation of cervical neoplastic cells infected with human papillomavirus 16. Int J Gynecol Cancer. 26:176–183. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Deng J, Wang Y, Zhang S and Chen L: A novel long noncoding RNA located on the antisense strand of MAL promotes the invasion and metastasis of oral squamous cell carcinoma. Arch Oral Biol. 155:1057902023. View Article : Google Scholar : PubMed/NCBI | |
|
Meršaková S, Holubeková V, Grendár M, Višňovský J, Ňachajová M, Kalman M, Kúdela E, Žúbor P, Bielik T, Lasabová Z and Danko J: Methylation of CADM1 and MAL together with HPV status in cytological cervical specimens serves an important role in the progression of cervical intraepithelial neoplasia. Oncol Lett. 16:7166–7174. 2018.PubMed/NCBI | |
|
Ondič O, Němcová J, Alaghehbandan R, Černá K, Gomolčáková B, Kinkorová-Luňáčková I, Chytra J, Šidlová H, Májek O and Bouda J: The detection of DNA methylation of tumour suppressor genes in cervical high-grade squamous intraepithelial lesion: A prospective cytological-histological correlation study of 70 cases. Cytopathology. 30:426–431. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Berchuck A, Iversen ES, Luo J, Clarke JP, Horne H, Levine DA, Boyd J, Alonso MA, Secord AA, Bernardini MQ, et al: Microarray analysis of early stage serous ovarian cancers shows profiles predictive of favorable outcome. Clin Cancer Res. 15:2448–2455. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Kulbe H, Otto R, Darb-Esfahani S, Lammert H, Abobaker S, Welsch G, Chekerov R, Schäfer R, Dragun D, Hummel M, et al: Discovery and validation of novel biomarkers for detection of epithelial ovarian cancer. Cells. 8:7132019. View Article : Google Scholar : PubMed/NCBI | |
|
Vasiljević N, Ahmad AS, Thorat MA, Fisher G, Berney DM, Møller H, Foster CS, Cuzick J and Lorincz AT: DNA methylation gene-based models indicating independent poor outcome in prostate cancer. BMC Cancer. 14:6552014. View Article : Google Scholar : PubMed/NCBI | |
|
Ahmad AS, Vasiljević N, Carter P, Berney DM, Møller H, Foster CS, Cuzick J and Lorincz AT: A novel DNA methylation score accurately predicts death from prostate cancer in men with low to intermediate clinical risk factors. Oncotarget. 7:71833–71840. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Choi P and Chen C: Genetic expression profiles and biologic pathway alterations in head and neck squamous cell carcinoma. Cancer. 104:1113–1128. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Riva G, Biolatti M, Pecorari G, Dell'Oste V and Landolfo S: PYHIN proteins and HPV: Role in the pathogenesis of head and neck squamous cell carcinoma. Microorganisms. 8:142019. View Article : Google Scholar : PubMed/NCBI | |
|
Misawa K, Imai A, Matsui H, Kanai A, Misawa Y, Mochizuki D, Mima M, Yamada S, Kurokawa T, Nakagawa T and Mineta H: Identification of novel methylation markers in HPV-associated oropharyngeal cancer: Genome-wide discovery, tissue verification and validation testing in ctDNA. Oncogene. 39:4741–4755. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Jiang Y, Chen Y, Gao L, Ye Q and Alonso MA: Expression pattern of MAL in normal epithelial cells, benign tumor, and squamous cell carcinoma of larynx. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 23:451–453. 2009.(In Chinese). PubMed/NCBI | |
|
Wilson SH, Bailey AM, Nourse CR, Mattei MG and Byrne JA: Identification of MAL2, a novel member of the mal proteolipid family, though interactions with TPD52-like proteins in the yeast two-hybrid system. Genomics. 76:81–88. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
In JG, Striz AC, Bernad A and Tuma PL: Serine/threonine kinase 16 and MAL2 regulate constitutive secretion of soluble cargo in hepatic cells. Biochem J. 463:201–213. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Marazuela M, Martín-Belmonte F, García-López MA, Aranda JF, de Marco MC and Alonso MA: Expression and distribution of MAL2, an essential element of the machinery for basolateral-to-apical transcytosis, in human thyroid epithelial cells. Endocrinology. 145:1011–1016. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Llorente A, de Marco MC and Alonso MA: Caveolin-1 and MAL are located on prostasomes secreted by the prostate cancer PC-3 cell line. J Cell Sci. 117:5343–5351. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Fanayan S, Shehata M, Agterof AP, McGuckin MA, Alonso MA and Byrne JA: Mucin 1 (MUC1) is a novel partner for MAL2 in breast carcinoma cells. BMC Cell Biol. 10:72009. View Article : Google Scholar : PubMed/NCBI | |
|
In JG and Tuma PL: MAL2 selectively regulates polymeric IgA receptor delivery from the Golgi to the plasma membrane in WIF-B cells. Traffic. 11:1056–1066. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
López-Coral A, Del Vecchio GJ, Chahine JJ, Kallakury BV and Tuma PL: MAL2-induced actin-based protrusion formation is anti-oncogenic in hepatocellular carcinoma. Cancers (Basel). 12:4222020. View Article : Google Scholar : PubMed/NCBI | |
|
Farazi PA and DePinho RA: The genetic and environmental basis of hepatocellular carcinoma. Discov Med. 6:182–186. 2006.PubMed/NCBI | |
|
Bhandari A, Shen Y, Sindan N, Xia E, Gautam B, Lv S and Zhang X: MAL2 promotes proliferation, migration, and invasion through regulating epithelial-mesenchymal transition in breast cancer cell lines. Biochem Biophys Res Commun. 504:434–439. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
An L, Gong H, Yu X, Zhang W, Liu X, Yang X, Shu L, Liu J and Yang L: Downregulation of MAL2 inhibits breast cancer progression through regulating β-catenin/c-Myc axis. Cancer Cell Int. 23:1442023. View Article : Google Scholar : PubMed/NCBI | |
|
Zhong Y, Zhuang Z, Mo P, Shang Q, Lin M, Gong J, Huang J, Mo H and Huang M: Overexpression of MAL2 correlates with immune infiltration and poor prognosis in breast cancer. Evid Based Complement Alternat Med. 2021:55578732021. View Article : Google Scholar : PubMed/NCBI | |
|
Yuan J, Jiang X, Lan H, Zhang X, Ding T, Yang F, Zeng D, Yong J, Niu B and Xiao S: Multi-omics analysis of the therapeutic value of MAL2 based on data mining in human cancers. Front Cell Dev Biol. 9:7366492022. View Article : Google Scholar : PubMed/NCBI | |
|
Jeong J, Shin JH, Li W, Hong JY, Lim J, Hwang JY, Chung JJ, Yan Q, Liu Y, Choi J and Wysolmerski J: MAL2 mediates the formation of stable HER2 signaling complexes within lipid raft-rich membrane protrusions in breast cancer cells. Cell Rep. 37:1101602021. View Article : Google Scholar : PubMed/NCBI | |
|
Dersh D and Yewdell JW: Immune MAL2-practice: Breast cancer immunoevasion via MHC class I degradation. J Clin Invest. 131:e1443442021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhu X, Bu J, Zhu T and Jiang Y: Targeting KK-LC-1 inhibits malignant biological behaviors of triple-negative breast cancer. J Transl Med. 21:1842023. View Article : Google Scholar : PubMed/NCBI | |
|
Eguchi D, Ohuchida K, Kozono S, Ikenaga N, Shindo K, Cui L, Fujiwara K, Akagawa S, Ohtsuka T, Takahata S, et al: MAL2 expression predicts distant metastasis and short survival in pancreatic cancer. Surgery. 154:573–582. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Xiong F, Wu GH, Wang B and Chen YJ: Plastin-3 is a diagnostic and prognostic marker for pancreatic adenocarcinoma and distinguishes from diffuse large B-cell lymphoma. Cancer Cell Int. 21:4112021. View Article : Google Scholar : PubMed/NCBI | |
|
Chen Y, Zheng B, Robbins DH, Lewin DN, Mikhitarian K, Graham A, Rumpp L, Glenn T, Gillanders WE, Cole DJ, et al: Accurate discrimination of pancreatic ductal adenocarcinoma and chronic pancreatitis using multimarker expression data and samples obtained by minimally invasive fine needle aspiration. Int J Cancer. 120:1511–1517. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Gao X, Chen Z, Li A, Zhang X and Cai X: MiR-129 regulates growth and invasion by targeting MAL2 in papillary thyroid carcinoma. Biomed Pharmacother. 105:1072–1078. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Li J, Li Y, Liu H, Liu Y and Cui B: The four-transmembrane protein MAL2 and tumor protein D52 (TPD52) are highly expressed in colorectal cancer and correlated with poor prognosis. PLoS One. 12:e01785152017. View Article : Google Scholar : PubMed/NCBI | |
|
Wang K, Yang Y, Zheng S and Hu W: Association mining identifies MAL2 as a novel tumor suppressor in colorectal cancer. Onco Targets Ther. 15:761–769. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Shao B, Fu X, Li X, Li Y and Gan N: RP11-284F21.9 promotes oral squamous cell carcinoma development via the miR-383-5p/MAL2 axis. J Oral Pathol Med. 49:21–29. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Dasgupta S, Tripathi PK, Qin H, Bhattacharya-Chatterjee M, Valentino J and Chatterjee SK: Identification of molecular targets for immunotherapy of patients with head and neck squamous cell carcinoma. Oral Oncol. 42:306–316. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Weis VG, Petersen CP, Mills JC, Tuma PL, Whitehead RH and Goldenring JR: Establishment of novel in vitro mouse chief cell and SPEM cultures identifies MAL2 as a marker of metaplasia in the stomach. Am J Physiol Gastrointest Liver Physiol. 307:G777–G792. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Guo JU, Agarwal V, Guo H and Bartel DP: Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI | |
|
Smolarz B, Zadrożna-Nowak A and Romanowicz H: The role of lncRNA in the development of tumors, including breast cancer. Int J Mol Sci. 22:84272021. View Article : Google Scholar : PubMed/NCBI | |
|
He J, Yan H, Wei S and Chen G: LncRNA ST8SIA6-AS1 promotes cholangiocarcinoma progression by suppressing the miR-145-5p/MAL2 axis. Onco Targets Ther. 14:3209–3223. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang C, Xu L, Li X, Chen Y, Shi T and Wang Q: LINC00460 facilitates cell proliferation and inhibits ferroptosis in breast cancer through the miR-320a/MAL2 axis. Technol Cancer Res Treat. 22:153303382311643592023. View Article : Google Scholar : PubMed/NCBI | |
|
Le Guelte A and Macara IG: Plasmolipin-a new player in endocytosis and epithelial development. EMBO J. 34:1147–1148. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Rodríguez-Fraticelli AE, Bagwell J, Bosch-Fortea M, Boncompain G, Reglero-Real N, García-León MJ, Andrés G, Toribio ML, Alonso MA, Millán J, et al: Developmental regulation of apical endocytosis controls epithelial patterning in vertebrate tubular organs. Nat Cell Biol. 17:241–250. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
You J, Corley SM, Wen L, Hodge C, Höllhumer R, Madigan MC, Wilkins MR and Sutton G: RNA-Seq analysis and comparison of corneal epithelium in keratoconus and myopia patients. Sci Rep. 8:3892018. View Article : Google Scholar : PubMed/NCBI | |
|
Yaffe Y, Hugger I, Yassaf IN, Shepshelovitch J, Sklan EH, Elkabetz Y, Yeheskel A, Pasmanik-Chor M, Benzing C, Macmillan A, et al: The myelin proteolipid plasmolipin forms oligomers and induces liquid-ordered membranes in the Golgi complex. J Cell Sci. 128:2293–2302. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Fischer I, Cochary EF, Konola JT and Romano-Clark G: Expression of plasmolipin in oligodendrocytes. J Neurosci Res. 28:81–89. 1991. View Article : Google Scholar : PubMed/NCBI | |
|
Fischer I, Durrie R and Sapirstein VS: Plasmolipin: The other myelin proteolipid. A review of studies on its structure, expression, and function. Neurochem Res. 19:959–966. 1994. View Article : Google Scholar : PubMed/NCBI | |
|
Shea TB, Fischer I and Sapirstein V: Expression of a plasma membrane proteolipid during differentiation of neuronal and glial cells in primary culture. J Neurochem. 47:697–706. 1986. View Article : Google Scholar : PubMed/NCBI | |
|
Hasse B, Bosse F and Müller HW: Proteins of peripheral myelin are associated with glycosphingolipid/cholesterol-enriched membranes. J Neurosci Res. 69:227–232. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Fredriksson K, Van Itallie CM, Aponte A, Gucek M, Tietgens AJ and Anderson JM: Proteomic analysis of proteins surrounding occludin and claudin-4 reveals their proximity to signaling and trafficking networks. PLoS One. 10:e01170742015. View Article : Google Scholar : PubMed/NCBI | |
|
Sapirstein VS, Nolan C, Stern R, Ciocci M and Masur SK: Identification of the plasma membrane proteolipid protein as a constituent of brain coated vesicles and synaptic plasma membrane. J Neurochem. 51:925–933. 1988. View Article : Google Scholar : PubMed/NCBI | |
|
Sapirstein VS, Nolan CE, Stern R, Gray-Board G and Beard ME: Identification of plasmolipin as a major constituent of white matter clathrin-coated vesicles. J Neurochem. 58:1372–1378. 1992. View Article : Google Scholar : PubMed/NCBI | |
|
Azzaz F, Mazzarino M, Chahinian H, Yahi N, Scala CD and Fantini J: Structure of the myelin sheath proteolipid plasmolipin (PLLP) in a ganglioside-containing lipid raft. Front Biosci (Landmark Ed). 28:1572023. View Article : Google Scholar : PubMed/NCBI | |
|
Gillen C, Gleichmann M, Greiner-Petter R, Zoidl G, Kupfer S, Bosse F, Auer J and Müller HW: Full-lenth cloning, expression and cellular localization of rat plasmolipin mRNA, a proteolipid of PNS and CNS. Eur J Neurosci. 8:405–414. 1996. View Article : Google Scholar : PubMed/NCBI | |
|
Fischer I and Sapirstein VS: Molecular cloning of plasmolipin. Characterization of a novel proteolipid restricted to brain and kidney. J Biol Chem. 269:24912–24919. 1994. View Article : Google Scholar : PubMed/NCBI | |
|
Bosse F, Hasse B, Pippirs U, Greiner-Petter R and Müller HW: Proteolipid plasmolipin: Localization in polarized cells, regulated expression and lipid raft association in CNS and PNS myelin. J Neurochem. 86:508–518. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Dannaeus K, Bessonova M and Jönsson JI: Characterization of the mouse myeloid-associated differentiation marker (MYADM) gene: Promoter analysis and protein localization. Mol Biol Rep. 32:149–157. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Takematsu E, Spencer A, Auster J, Chen PC, Graham A, Martin P and Baker AB: Genome wide analysis of gene expression changes in skin from patients with type 2 diabetes. PLoS One. 15:e02252672020. View Article : Google Scholar : PubMed/NCBI | |
|
Zheng P, Sun S, Wang J, Cheng ZJ, Lei KC, Xue M, Zhang T, Huang H, Zhang XD and Sun B: Integrative omics analysis identifies biomarkers of idiopathic pulmonary fibrosis. Cell Mol Life Sci. 79:662022. View Article : Google Scholar : PubMed/NCBI | |
|
Lautner-Rieske A, Thiebe R and Zachau HG: Searching for non-V kappa transcripts from the human immunoglobulin kappa locus. Gene. 159:199–202. 1995. View Article : Google Scholar : PubMed/NCBI | |
|
de Marco MC, Kremer L, Albar JP, Martinez-Menarguez JA, Ballesta J, Garcia-Lopez MA, Marazuela M, Puertollano R and Alonso MA: BENE, a novel raft-associated protein of the MAL proteolipid family, interacts with caveolin-1 in human endothelial-like ECV304 cells. J Biol Chem. 276:23009–23017. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Rubio-Ramos A, Bernabé-Rubio M, Labat-de-Hoz L, Casares-Arias J, Kremer L, Correas I and Alonso MA: MALL, a membrane-tetra-spanning proteolipid overexpressed in cancer, is present in membraneless nuclear biomolecular condensates. Cell Mol Life Sci. 79:2362022. View Article : Google Scholar : PubMed/NCBI | |
|
Kim K, Park U, Wang J, Lee J, Park S, Kim S, Choi D, Kim C and Park J: Gene profiling of colonic serrated adenomas by using oligonucleotide microarray. Int J Colorectal Dis. 23:569–580. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Kim HA, Kim KH and Lee RA: Expression of caveolin-1 is correlated with Akt-1 in colorectal cancer tissues. Exp Mol Pathol. 80:165–170. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Wang X, Fan J, Yu F, Cui F, Sun X, Zhong L, Yan D, Zhou C, Deng G, Wang B, et al: Decreased MALL expression negatively impacts colorectal cancer patient survival. Oncotarget. 7:22911–22927. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Li DX, Yu QX, Zeng CX, Ye LX, Guo YQ, Liu JF, Zheng HH, Feng D and Wei W: A novel endothelial-related prognostic index by integrating single-cell and bulk RNA sequencing data for patients with kidney renal clear cell carcinoma. Front Genet. 14:10964912023. View Article : Google Scholar : PubMed/NCBI | |
|
Aranda JF, Reglero-Real N, Marcos-Ramiro B, Ruiz-Sáenz A, Fernández-Martín L, Bernabé-Rubio M, Kremer L, Ridley AJ, Correas I, Alonso MA and Millán J: MYADM controls endothelial barrier function through ERM-dependent regulation of ICAM-1 expression. Mol Biol Cell. 24:483–494. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Q, Li N, Wang X, Shen J, Hong X, Yu H, Zhang Y, Wan T, Zhang L, Wang J and Cao X: Membrane protein hMYADM preferentially expressed in myeloid cells is up-regulated during differentiation of stem cells and myeloid leukemia cells. Life Sci. 80:420–429. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Cui W, Yu L, He H, Chu Y, Gao J, Wan B, Tang L and Zhao S: Cloning of human myeloid-associated differentiation marker (MYADM) gene whose expression was up-regulated in NB4 cells induced by all-trans retinoic acid. Mol Biol Rep. 28:123–138. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Pettersson M, Dannaeus K, Nilsson K and Jönsson JI: Isolation of MYADM, a novel hematopoietic-associated marker gene expressed in multipotent progenitor cells and up-regulated during myeloid differentiation. J Leukoc Biol. 67:423–431. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Dy ABC, Langlais PR, Barker NK, Addison KJ, Tanyaratsrisakul S, Boitano S, Christenson SA, Kraft M, Meyers D, Bleecker ER, et al: Myeloid-associated differentiation marker is a novel SP-A-associated transmembrane protein whose expression on airway epithelial cells correlates with asthma severity. Sci Rep. 11:233922021. View Article : Google Scholar : PubMed/NCBI | |
|
Sun L, Lin P, Chen Y, Yu H, Ren S, Wang J, Zhao L and Du G: miR-182-3p/Myadm contribute to pulmonary artery hypertension vascular remodeling via a KLF4/p21-dependent mechanism. Theranostics. 10:5581–5599. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
de Wit NJ, Rijntjes J, Diepstra JH, van Kuppevelt TH, Weidle UH, Ruiter DJ and van Muijen GN: Analysis of differential gene expression in human melanocytic tumour lesions by custom made oligonucleotide arrays. Br J Cancer. 92:2249–2261. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Megger DA, Naboulsi W, Meyer HE and Sitek B: Proteome analyses of hepatocellular carcinoma. J Clin Transl Hepatol. 2:23–30. 2014.PubMed/NCBI | |
|
Zhou M, Chen Y, Gu X and Wang C: A comprehensive bioinformatic analysis for identification of myeloid-associated differentiation marker as a potential negative prognostic biomarker in non-small-cell lung cancer. Pathol Oncol Res. 28:16105042022. View Article : Google Scholar : PubMed/NCBI | |
|
Wang X, Dou X, Ren X, Rong Z, Sun L, Deng Y, Chen P and Li Z: A ductal-cell-related risk model integrating single-cell and bulk sequencing data predicts the prognosis of patients with pancreatic adenocarcinoma. Front Genet. 12:7636362021. View Article : Google Scholar : PubMed/NCBI | |
|
Papasotiriou I, Apostolou P, Ntanovasilis DA, Parsonidis P, Osmonov D and Jünemann KP: Study and detection of potential markers for predicting metastasis into lymph nodes in prostate cancer. Biomark Med. 14:1317–1327. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Echevarria MI, Awasthi S, Cheng CH, Berglund AE, Rounbehler RJ, Gerke TA, Takhar M, Davicioni E, Klein EA, Freedland SJ, et al: African American specific gene panel predictive of poor prostate cancer outcome. J Urol. 202:247–255. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang B, Ren Z, Zheng H, Lin M, Chen G, Luo OJ and Zhu G: CRISPR activation screening in a mouse model for drivers of hepatocellular carcinoma growth and metastasis. iScience. 26:1060992023. View Article : Google Scholar : PubMed/NCBI | |
|
Li D, Jin C, Yin C, Zhang Y, Pang B, Tian L, Han W, Ma D and Wang Y: An alternative splice form of CMTM8 induces apoptosis. Int J Biochem Cell Biol. 39:2107–2119. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Jin C, Ding P, Wang Y and Ma D: Regulation of EGF receptor signaling by the MARVEL domain-containing protein CKLFSF8. FEBS Lett. 579:6375–6382. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Jin C, Wang Y, Han W, Zhang Y, He Q, Li D, Yin C, Tian L, Liu D, Song Q and Ma D: CMTM8 induces caspase-dependent and -independent apoptosis through a mitochondria-mediated pathway. J Cell Physiol. 211:112–120. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Yan M, Zhu X, Qiao H, Zhang H, Xie W and Cai J: Downregulated CMTM8 correlates with poor prognosis in gastric cancer patients. DNA Cell Biol. 40:791–797. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Wu K, Li X, Gu H, Yang Q, Liu Y and Wang L: Research advances in CKLF-like MARVEL transmembrane domain-containing family in non-small cell lung cancer. Int J Biol Sci. 15:2576–2583. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Hu H, Chen JW, Xu KX, Wang D, Wang Y, Wang GW, Zhang SY and Wang XF: Expressions of CMTM8 and E-cadherin in primary and metastatic clear cell renal cell carcinoma. Beijing Da Xue Xue Bao Yi Xue Ban. 45:537–541. 2013.(In Chinese). PubMed/NCBI | |
|
Zhang W, Mendoza MC, Pei X, Ilter D, Mahoney SJ, Zhang Y, Ma D, Blenis J and Wang Y: Down-regulation of CMTM8 induces epithelial-to-mesenchymal transition-like changes via c-MET/extracellular signal-regulated kinase (ERK) signaling. J Biol Chem. 287:11850–11858. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Zeng X, Ma X, Guo H, Wei L, Zhang Y, Sun C, Han N, Sun S and Zhang N: MicroRNA-582-5p promotes triple-negative breast cancer invasion and metastasis by antagonizing CMTM8. Bioengineered. 12:10126–10135. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Both J, Krijgsman O, Bras J, Schaap GR, Baas F, Ylstra B and Hulsebos TJ: Focal chromosomal copy number aberrations identify CMTM8 and GPR177 as new candidate driver genes in osteosarcoma. PLoS One. 9:e1158352014. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang S, Pei X, Hu H, Zhang W, Mo X, Song Q, Zhang Y, Xu K, Wang Y and Na Y: Functional characterization of the tumor suppressor CMTM8 and its association with prognosis in bladder cancer. Tumour Biol. 37:6217–6225. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Gao D, Hu H, Wang Y, Yu W, Zhou J, Wang X, Wang W, Zhou C and Xu K: CMTM8 inhibits the carcinogenesis and progression of bladder cancer. Oncol Rep. 34:2853–2863. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Kang N, Xie X, Zhou X, Wang Y, Chen S, Qi R, Liu T and Jiang H: Identification and validation of EMT-immune-related prognostic biomarkers CDKN2A, CMTM8 and ILK in colon cancer. BMC Gastroenterol. 22:1902022. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang M, Wang J, Yue H and Zhang L: Identification of prognostic biomarkers in the CMTM family genes of human ovarian cancer through bioinformatics analysis and experimental verification. Front Genet. 13:9183192022. View Article : Google Scholar : PubMed/NCBI | |
|
Shi W, Zhang C, Ning Z, Hua Y, Li Y, Chen L, Liu L, Chen Z and Meng Z: CMTM8 as an LPA1-associated partner mediates lysophosphatidic acid-induced pancreatic cancer metastasis. Ann Transl Med. 9:422021. View Article : Google Scholar : PubMed/NCBI | |
|
Liu Y, Chew MH, Tham CK, Tang CL, Ong SY and Zhao Y: Methylation of serum SST gene is an independent prognostic marker in colorectal cancer. Am J Cancer Res. 6:2098–2108. 2016.PubMed/NCBI | |
|
Guerrero-Preston R, Hadar T, Ostrow KL, Soudry E, Echenique M, Ili-Gangas C, Pérez G, Perez J, Brebi-Mieville P, Deschamps J, et al: Differential promoter methylation of kinesin family member 1a in plasma is associated with breast cancer and DNA repair capacity. Oncol Rep. 32:505–512. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Hentschel AE, Nieuwenhuijzen JA, Bosschieter J, Splunter APV, Lissenberg-Witte BI, Voorn JPV, Segerink LI, Moorselaar RJAV and Steenbergen RDM: Comparative analysis of urine fractions for optimal bladder cancer detection using DNA methylation markers. Cancers (Basel). 12:8592020. View Article : Google Scholar : PubMed/NCBI | |
|
Lind GE, Danielsen SA, Ahlquist T, Merok MA, Andresen K, Skotheim RI, Hektoen M, Rognum TO, Meling GI, Hoff G, et al: Identification of an epigenetic biomarker panel with high sensitivity and specificity for colorectal cancer and adenomas. Mol Cancer. 10:852011. View Article : Google Scholar : PubMed/NCBI | |
|
Obermayr E, Sanchez-Cabo F, Tea MK, Singer CF, Krainer M, Fischer MB, Sehouli J, Reinthaller A, Horvat R, Heinze G, et al: Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients. BMC Cancer. 10:6662010. View Article : Google Scholar : PubMed/NCBI | |
|
Obermayr E, Maritschnegg E, Agreiter C, Pecha N, Speiser P, Helmy-Bader S, Danzinger S, Krainer M, Singer C and Zeillinger R: Efficient leukocyte depletion by a novel microfluidic platform enables the molecular detection and characterization of circulating tumor cells. Oncotarget. 9:812–823. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Ding W, Li Z, Wang C, Dai J, Ruan G and Tu C: Anthracycline versus nonanthracycline adjuvant therapy for early breast cancer: A systematic review and meta-analysis. Medicine (Baltimore). 97:e129082018. View Article : Google Scholar : PubMed/NCBI | |
|
Willson ML, Burke L, Ferguson T, Ghersi D, Nowak AK and Wilcken N: Taxanes for adjuvant treatment of early breast cancer. Cochrane Database Syst Rev. 9:CD0044212019.PubMed/NCBI | |
|
Arumugam T, Ramachandran V, Fournier KF, Wang H, Marquis L, Abbruzzese JL, Gallick GE, Logsdon CD, McConkey DJ and Choi W: Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 69:5820–5828. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Bracker TU, Sommer A, Fichtner I, Faus H, Haendler B and Hess-Stumpp H: Efficacy of MS-275, a selective inhibitor of class I histone deacetylases, in human colon cancer models. Int J Oncol. 35:909–920. 2009.PubMed/NCBI |