Lidocaine attenuates TMZ resistance and inhibits cell migration by modulating the MET pathway in glioblastoma cells
- Authors:
- Published online on: April 8, 2024 https://doi.org/10.3892/or.2024.8731
- Article Number: 72
-
Copyright: © Chen et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Glioblastoma multiforme (GBM) is a highly malignant and aggressive type of astrocytic tumor. The World Health Organization (WHO) has classified GBM as a grade IV astrocytoma. GBM is the most common form of brain tumor, accounting for 16% of all primary brain tumors and 54% of all gliomas (1,2). Currently, the primary clinical approach for managing GBM involves extensive surgical resection to remove as much of the tumor as possible, followed by a combination therapy of radiotherapy and temozolomide (TMZ) chemotherapy (3). Despite receiving the standard treatment regimen, GBM patients have a median survival of only 15 months. This underscores the pressing need for developing novel treatments to improve the prognosis of GBM patients (4). Currently, there are several clinical online databases available for researchers to utilize for the identification of new therapeutic targets. Using these databases, it was found that the hepatocyte growth factor (HGF)/mesenchymal-epithelial transition factor (MET) pathway has potential therapeutic targets. The human MET receptor gene is located on chromosome 7q31 and encodes a 50-kD α subunit and a 140-kD β subunit. These subunits combine to form a 190-kD heterodimeric transmembrane receptor. The only known ligand for MET is hepatocyte growth factor (HGF), also referred to as scatter factor (SF), which is encoded by a gene found on chromosome 7q21.1 (5). MET signaling is initiated when HGF binds to the MET receptor, which is essential for embryonic development and tissue regeneration (6). Abnormal HGF/MET signaling has been implicated in the progression of brain tumors (7). Increased expression of HGF and MET in GBM tissue has been found to be strongly associated with poorer prognoses for patients, therefore it can be used as an independent predictor of patient survival (8–13). Moreover, drugs that target HGF/MET are expected to become a therapeutic option for GBM patients in the future (14,15).
Lidocaine, an amide-type local anesthetic, is widely used to manage several acute or chronic pain disorders, such as inflammation, nociceptive pain and neuropathic pain (16). Recently, lidocaine has been found to have a significant anticancer effect in various types of cancer. It can reduce the progression and recurrence of cancer and improve the survival rate of patients (17). Numerous studies indicated that lidocaine can suppress malignant cell behavior, including growth, proliferation, migration and invasion, as well as induce cell apoptosis in a wide range of cancer cells (18–24). It also has a sensitizing effect on clinical chemotherapeutic drugs (25–27). Chemotherapy is an important method for treating various cancers, but some patients develop drug resistance over time, limiting the efficacy of chemotherapy and ultimately leading to the patient's death. The latest research revealed that lidocaine can alleviate chemotherapy resistance (28,29).
There is increasing evidence suggesting that lidocaine can suppress cell proliferation, migration, invasion and promote apoptosis in glioma cells (30–33). A recent study also demonstrated that lidocaine can hinder the survival and self-renewal abilities of GBM stem cells (34). These findings indicated that lidocaine could potentially serve as a therapeutic option for GBM, although its specific mechanism of action remains largely unknown. Mechanistically, recent research suggests that lidocaine can significantly reduce MET activation (35). However, whether lidocaine can inhibit the HGF/MET signaling in GBM cells has not been reported yet. Therefore, the present study primarily aimed to investigate whether lidocaine can regulate HGF/MET-related pathways in GBM cells and further elucidate the anticancer effect of lidocaine.
Materials and methods
Public database analysis
The mRNA expression levels of HGF and MET were analyzed in human tissue samples from normal brain, brain cancer, glioma, and GBM using the R2 Genomics Analysis and Visualization Platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). The results were presented as median and interquartile range. Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/index.html), an online web server based on The Cancer Genome Atlas (TCGA) and GTEx data, was also used to analyze the mRNA expression levels of HGF and MET in GBM tissues and normal tissues. The GBM dataset was selected for analysis, with |Log2FC| cutoff set to 1 and the P-value cutoff set to 0.01. Whisker plots were generated to show the relative RNA expression levels of HGF and MET in GBM tissues (n=163) and normal tissues (n=207). In addition, we utilized The Cancer Proteome Atlas (TCPA) database (http://tcpaportal.org/tcpa/) to examine the protein expression levels of MET pY1235 and MET in GBM samples. To determine the potential relationship between the expression of HGF/MET and patient survival, an analysis of the R2 database was conducted to evaluate the association between their expression levels and clinical prognosis in brain tumors and gliomas. The Kaplan-Meier curve was generated using the following datasets: Tumor Brain-Madhavan-550-MAS5.0-u133p2 (HGF, 210998_s_at; MET, 203510_at); Tumor Glioma-French-284-AS5.0-u133p2 (HGF, 209960_at; MET, 203510_at); Tumor Brain Lower Grade Glioma (2022-v2)-tcga-532-tpm-gencode36 (HGF, ENSG00000019991.18; MET, ENSG00000105976.16); and Tumor Glioblastoma-TCGA-540-MAS5.0-u133a (HGF, 209960_at; MET, 203510_at). In the Glioma database (French, n=284), patient survival information is available for only 273 samples. In addition, the Kaplan-Meier survival plots were generated using the TCGA database to analyze the relationship between the protein/phosphoprotein expressions of MET and patient survival.
Chemicals and reagents
Lidocaine hydrochloride, obtained from Aspen Pharma Trading Ltd., was stored at 4°C until use. Additional chemicals, including TMZ, DMSO, MTT, and other reagents, were purchased from MilliporeSigma. The primary antibodies employed in the present study included HGF (cat. no. ab83760), MET (cat. no. ab216574), phosphorylated (p)-MET (cat. no. ab5662), and cleaved PARP (cat. no. ab32064), all of which were purchased from Abcam. The antibody for α-tubulin (cat. no. 05-829) was acquired from EMD Millipore. The secondary antibody HRP-conjugated goat anti-rabbit IgG was obtained from Leadgene Biomedical, Inc. (cat. no. 20202), while the secondary antibody HRP-conjugated goat anti-mouse IgG was procured from Jackson ImmunoResearch Laboratories, Inc. (cat. no. 115-035-003).
Cell culture
The U87MG (glioblastoma of unknown origin) and DBTRG-05MG cell lines were obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin/streptomycin at 37°C in a humidified 5% CO2 atmosphere. All cell culture reagents were purchased from Gibco; Thermo Fisher Scientific, Inc. RNA sequencing was conducted to validate the U87MG cell line, as evidenced by the data provided in the supplementary material. To investigate the mechanism of TMZ resistance in GBM, the cells were gradually exposed to increasing doses of TMZ (ranging from 15.625 to 250 µM) over a period of 6 months. Once the cells were able to survive in the drug, they were considered TMZ-resistant and designated as U87MGR cells. To analyze secreted proteins, 2×106 cells were seeded into a 10-cm dish and cultured until they reached ~80% confluency. The cells were then washed twice with 1X PBS to completely remove FBS. Subsequently, the cells were cultured in serum-free DMEM for 48 h, and the supernatant was collected for further analysis. To further determine the relationship between HGF and drug resistance, U87MG cells were pre-treated with conditioned media (CM) from both U87MG and U87MGR cells. Additionally, a portion of CM from U87MGR cells was pre-treated with HGF-neutralizing antibody (1:500) for 1 h before administering to U87MG cells. Then, the cells were treated with TMZ and lidocaine together for 48 h. All cell observation and imaging in the study were conducted using the Hamlet microscope.
Detection of apoptotic rate using flow cytometry
Cells were seeded at a density of 1.5×105 cells/well in 6-well plates and cultured overnight to allow adherence. After treatment with TMZ (0–1,000 µM) for 48 h, the cells were harvested, washed with cold PBS, and stained using the Annexin V FITC/PI Apoptosis Detection Kit (cat. no. 556547; BD Biosciences) in the dark at 25°C for 15 min. The apoptotic rate was then analyzed using flow cytometry. Flow cytometric analysis was conducted on a BD FACSCanto™ II flow cytometer (BD Biosciences), and data acquisition was carried out using FACSDiva software version 6.1 (BD Biosciences).
Western blotting
Cell lysates were prepared using RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate and 0.1% SDS) with a protease inhibitor cocktail. The concentration of total protein was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Inc.). Equal amounts (40 µg) of protein were separated on SDS-PAGE gels and transferred onto PVDF membranes (MilliporeSigma). The percentage of polyacrylamide used for the separation varied for different protein targets: HGF in conditioned medium: 12%, HGF in cytosol: 8%, pMET/MET: 8%, and cleaved PARP: 12%. The membranes were blocked with 5% skim milk dissolved in Tris-buffered saline (TBS) for 1 h at room temperature. Subsequently, the membranes were hybridized with specific primary antibodies overnight at 4°C. The primary antibodies used in western blotting were diluted as follows: α-tubulin at a ratio of 1:10,000, and HGF, p-MET, MET and cleaved PARP at a ratio of 1:1,000. Following several washes in TBS supplemented with 0.05% Tween-20 (TBST), the membranes were incubated with suitable HRP-conjugated secondary antibodies at room temperature for 1 h. The secondary antibodies used in western blotting were all diluted at a ratio of 1:5,000. After washing the membranes with TBST, the protein bands were visualized using ECL reagents (cat. no. 34580; Thermo Fisher Scientific, Inc.). α-tubulin was used as the loading control for normalization in western blotting. To analyze secreted proteins, cells were cultured in serum-free DMEM for 48 h, and the culture supernatants were then concentrated using a 3-kDa cut-off Amicon Ultra filter (Merck KGaA). Subsequently, the concentrated supernatants were subjected to protein quantification, and equal amounts of total protein were analyzed using western blotting. Due to the absence of specific markers for secretory proteins, equal amounts of total protein were loaded and confirmed using Ponceau staining for uniformity after protein quantification. The protein loading quantity was confirmed to be consistent through Ponceau S staining.
Cell cytotoxicity
To determine the effect of lidocaine and gene knockdown on TMZ sensitivity in GBM cells, the MTT assay was employed. In brief, 5×103 cells were seeded in 100 µl of culture medium per well on a 96-well plate and incubated overnight. The cells were then treated with lidocaine either alone or in combination with TMZ for indicated time periods. A total of 4 h prior to the completion of the experimental reaction, MTT reagent (5 mg/ml in PBS) was added to each well, and the cells were incubated for 4 h. The supernatant was subsequently removed, and 100 µl of DMSO was added to each well. The viability of cells was determined by measuring absorbance at 550 nm using a SpectraMax ABS Plus plate reader (Molecular Devices, LLC), and the background reading at 750 nm was subtracted. The results were expressed as a percentage, relative to untreated cells.
Colony formation assay
A total of 2×105 cells were seeded in 2 ml of culture medium per well on a 6-well plate and incubated overnight. Following treatment with lidocaine, TMZ, or a combination of both for 24 h, the cells were harvested and suspended in culture medium. Subsequently, the cells were seeded on a 6-well plate at a density of 5×103 cells per well and incubated in an incubator at 37°C with 5% CO2 for ~2 weeks until colonies formed (consisting of at least 50 cells). After washing with PBS, the cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Subsequently, they were stained with 0.1% crystal violet for 30 min, and the number of colonies was assessed using a light microscope and ImageJ software (version 2.1.0/1.5.3v; National Institutes of Health).
Cell proliferation assay
The cell proliferation was evaluated using the MTT assay. Briefly, 5,000 cells were seeded per well on 96-well plates and incubated for 0, 24, 48 and 72 h. MTT reagent (5 mg/ml in PBS) was added to each well, and the cells were incubated for 4 h. After incubation, the plates were centrifuged at 300 × g for 5 min. The supernatant was removed, and 100 µl of DMSO was added to each well. The absorbance was measured at 550 and 750 nm to subtract the background signal (750 nm). Cell proliferation was quantified by measuring the optical density.
Wound-healing assay
GBM cells were seeded on a 6-well plate until they reached 100% confluence. The cell monolayer was manually scratched using a 200 µl-sterile pipette tip to create a straight line. The wells were gently washed once with PBS to clean the field of view and images were captured as the controls. The cells were treated with serum-free medium containing lidocaine at 37°C for 24 h. At least five continuous fields per well were recorded before and 24 h after migration using a light microscope (magnification, ×200). The migration rate was calculated as follows: % of wound closure=100× (Scratching area at 0 h-Wound area at 24 h)/Scratching area at 0 h.
Transwell assay
A total of 1×105 cells were resuspended in 200 µl of serum-free culture medium and seeded onto the Transwell insert (8-µm pore size; Costar; Corning, Inc.). The lower chamber was filled with 300 µl of culture medium containing 10% FBS. The Transwell inserts were then placed in a 37°C incubator with 5% CO2 for 24 h. Afterwards, the Transwell inserts were fixed with 3.7% formaldehyde for 15 min and stained with 0.05% crystal violet for 30 min. The cells were washed with PBS, and any remaining stain on the upper side of the Transwell insert was removed with a cotton swab. The number of migratory cells on the lower side of the Transwell was observed under a light microscope at randomly selected different fields of view, images were captured, and cells were counted.
Gene knockdown
The third-generation lentiviral vectors utilized in the present study were sourced from the RNAi Core Facility at Academia Sinica located in Taipei, Taiwan. Using a transfection reagent (PolyJet; cat. no. SL100688; SignaGen Laboratories), a combination of 2 µg of specific short hairpin (sh)RNA, 2 µg of pCMVΔR8.91, and 0.2 µg of pMD plasmids was introduced into 293T cells (cat. no. CRL-3216; ATCC) to generate the recombinant lentiviruses. Transfection was carried out at 37°C overnight. Following this, a fresh medium replacement was performed, and the cells were further cultured for 24 to 72 h to collect lentiviral particles. GBM cells were infected with the lentiviral supernatant in the presence of polybrene (8 µg/ml). The lentiviral particles were then used to infect cells overnight at a multiplicity of infection (MOI) of 5. After transduction, puromycin (5 µg/ml) was added to the cells for 48 h to select stable cell lines. The lentiviral plasmids targeting HGF were TRCN0000047135 (shHGF#1, 5′-CCG-GCA-GAC-CAA-TGT-GCT-AAT-AGA-TCT-CGA-GAT-CTA-TTA-GCA-CAT-TGG-TC-TGT-TTT-T-3′) and TRCN0000047137 (shHGF#2, 5′-CCG-GGC-AAA-GAC-TAC-CCT-AAT-CAA-ACT-CGA-GTT-TGA-TTA-GGG-TAG-TCT-TTG-CTT-TTT-3′). The lentiviral plasmids targeting MET were TRCN0000040047 (shMET#1, 5′-CCG-GGC-CAG-CCT-GAA-TGA-TGA-CAT-TCT-CGA-GAA-TGT-CAT-CAT-TCA-GGC-TGG-CTT-TTT-G-3′) and TRCN0000009850 (shMET#2, 5′-CCG-GCA-GAA-TGT-CAT-TCT-ACA-TGA-GCT-CGA-GCT-CAT-GTA-GAA-TGA-CAT-TCT-GTT-TTT-G-3′). The control shRNA sequence used was TRC1. Scramble: 5′-CCG-GCC-TAA-GGT-TAA-GTC-GCC-CTC-GCT-CGA-GCG-AGG-GCG-ACT-TAA-CCT-TAG-GTT-TTT-3′. The sequence design was informed by the reference website (https://rnai.genmed.sinica.edu.tw/searchDatabase).
Reverse transcription-quantitative PCR (RT-qPCR)
After isolating total RNAs using REzol reagent (Protech Technology Enterprise CO., Ltd.) and reverse transcribing the mRNA to cDNA using a PrimeScript RT Reagent kit (Takara Bio, Inc.), the levels of HGF mRNA were detected using KAPA SYBR FAST qPCR Master Mix, according to the manufacturer's instructions. The MyGo PCR Detection System (IT-IS Life Science Ltd.) was used for detection. The following primers were used in the experiment: HGF forward, 5′-GAA-TGC-ATG-ACC-TGC-AAC-GG-3′ and reverse, 5′-TGT-CGG-GAT-ATC-TTT-CCG-GC-3′; and GAPDH forward, 5′-CAC-CCA-TGG-CAA-ATT-CCA-TGG-CA-3′ and reverse, 5′-TCT-AGA-CGG-CAG-GTC-AGG-TCC-ACC-3′. The RT-qPCR thermocycling conditions included an initial denaturation at 95°C for 180 sec, followed by 40 cycles with denaturation at 95°C for 10 sec, annealing at 60°C for 20 sec, and extension at 72°C for 30 sec. The relative level of gene expression was determined using the comparative Cq (2−ΔΔCq) method (36), with GAPDH used as the normalization reference.
Statistical analysis
The log-rank test was employed for Kaplan-Meier survival data analysis. The results are presented as the mean ± SEM, obtained from at a minimum of three separate experiments. The statistical analyses performed out using the Graph Pad Prism 6.0 software (Dotmatics). A comparison between two groups was performed using unpaired Student's t-test. When examining multiple variables, one-way analysis of variance with Bonferroni's post-hoc test was used. P<0.05 was considered to indicate a statistically significant difference.
Results
Using bioinformatics tools to analyze the association between the expression/activation of HGF/MET and its clinical significance in brain cancer patients
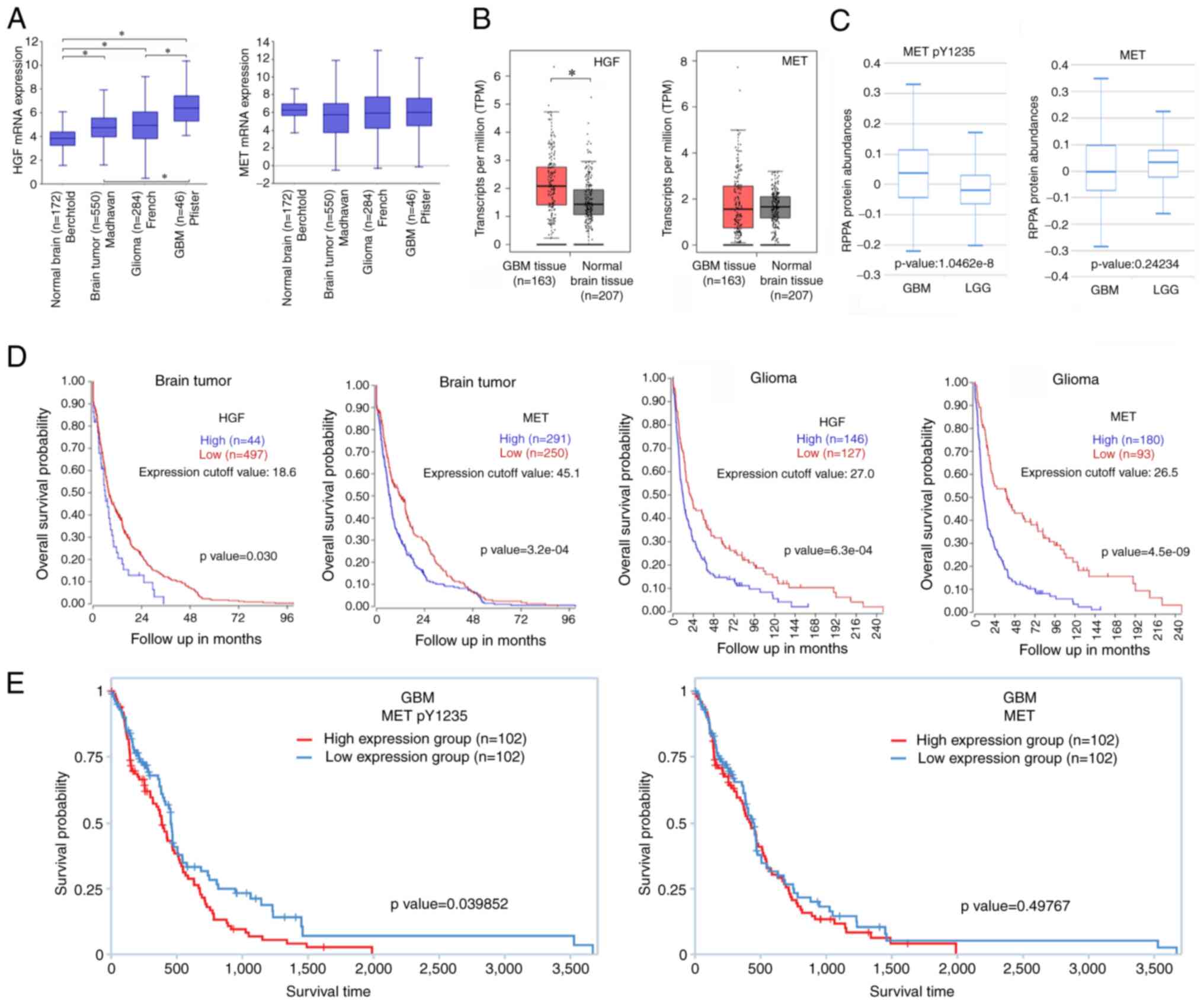
To understand the clinical significance of HGF and its receptor MET, publicly available online databases (R2 database) were used to compare the expression levels of these biomolecules in healthy brain tissues and tumor brain tissues. The results indicated that HGF expression in brain tumors (Madhavan), gliomas (French), and GBM (Pfister) was upregulated compared with normal brain tissues (Berchtold) (Fig. 1A). However, there was no significant difference in the expression levels of MET among the groups. In addition, GEPIA database was used to analyze the expression levels of HGF and MET in GBM. The mRNA expression levels of HGF in GBM were elevated compared with normal tissues, but there was not a significant difference in MET expression (Fig. 1B). As the mRNA levels of MET did not appear to be clinically related to the level of deterioration, TCPA database was utilized to analyze the protein expression levels of p-MET and MET in GBM and low-grade gliomas (LGG). Protein levels of p-MET were significantly higher in GBM compared with LGG (Fig. 1C). Next, the R2 database was utilized to obtain gene expression data in brain tumors and gliomas, allowing to examine the potential impact of HGF and MET expression on the overall survival of patients. The present analysis revealed that patients with tumors exhibiting elevated expression levels of HGF and MET experienced poorer survival outcomes (Fig. 1D). Information on the data obtained from the R2 database, including tumor type, patient age, sex distribution and sample size, are presented in Tables I and II. The results suggested that increased HGF and MET expression in brain tumors and gliomas may worsen malignancy, impacting patient survival. This emphasizes the imperative for an in-depth investigation into the roles of HGF and MET in GBM. Finally, the TCPA database was used to analyze the relationship between the levels of p-MET (Y1235)/MET and the survival probability of GBM patients. The results revealed that a higher phosphorylation level of MET was significantly associated with a poorer survival rate in GBM patients, while MET expression itself was not significantly associated to survival rate (Fig. 1E). Fig. 1A-C and 1E are all derived from the GBM database. However, it is important to clarify that the analysis in Fig. 1D actually comes from the brain tumor and glioma databases, not the GBM database. Interestingly, the results in Fig. 1D suggested that high expression of HGF and MET in brain tumors and gliomas may exacerbate their malignancy, thereby influencing patient survival rates. These findings indicated a crucial role for HGF/MET in brain cancer, yet in GBM, the actual activation of MET appears to be more significant than its expression level in influencing patient prognosis. Therefore, inhibiting abnormal HGF/MET axis activation may be a potential treatment strategy for GBM.
Activation of the HGF/MET pathway may be associated with TMZ resistance in GBM cells
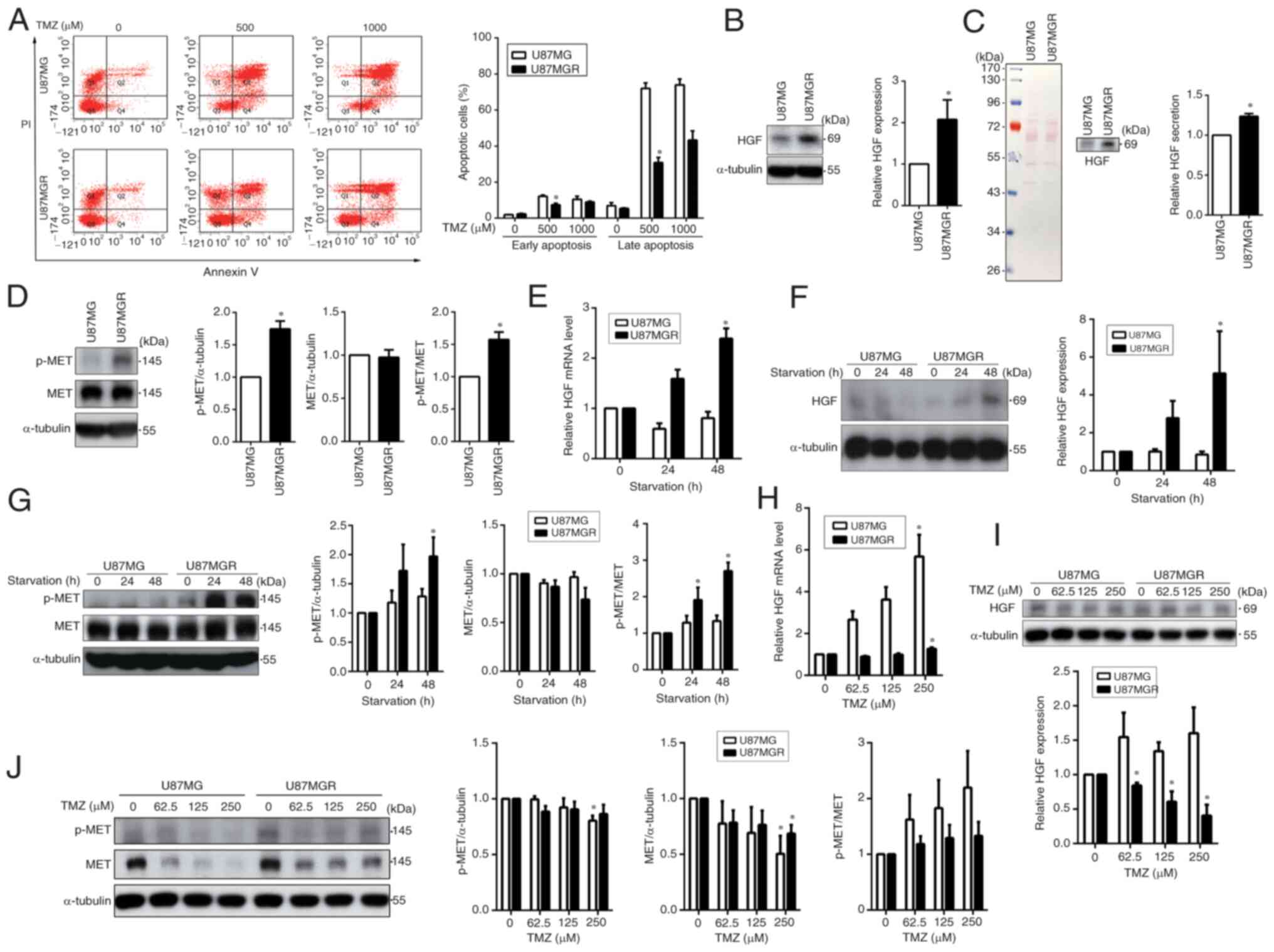
To investigate the relationship between the HGF/MET pathway and TMZ sensitivity in GBM cells, TMZ-resistant cells were established from U87MG cells, which are referred to as U87MGR. Flow cytometric analysis revealed that ~80% of parental cells underwent apoptosis when treated with 500 µM TMZ, whereas resistant cell lines exhibited an apoptotic rate of ~50% even after TMZ treatment at concentrations up to 1,000 µM, indicating that U87MGR cells were highly resistant to TMZ (Fig. 2A). Next, HGF expression was compared between the two cell lines and it was identified that the expression of HGF in U87MGR cells was higher than that in U87MG cells (Fig. 2B). Furthermore, cell culture supernatants were collected and concentrated for subsequent analysis using electrophoresis. Ponceau S staining showed that the total amount of loaded protein was consistent across the samples (Fig. 2C, left panel), and the results of western blotting revealed that HGF secretion by U87MGR cells was higher than that by U87MG cells (Fig. 2C, right panel). As high levels of secreted HGF can activate the MET pathway, the activation levels of MET in these cells was further analyzed. MET was highly activated in U87MGR cells compared with U87MG cells (Fig. 2D). During stressful conditions such as starvation or chemotherapy, cancer cells may activate certain signaling pathways that give them an advantage in survival (37). Therefore, U87MG and U87MGR cells were cultured in serum-free medium for 24 and 48 h and the changes in HGF expression and MET activation upon starvation were assessed. With increasing starvation time, TMZ-resistant cells exhibited elevated HGF levels, and this effect was particularly evident after 48 h of starvation (Fig. 2E and F). Notably, the levels of activation of the MET pathway induced by starvation in U87MGR cells were markedly higher compared with their parental cells (Fig. 2G). Next, U87MG and U87MGR cells were treated with increasing doses of TMZ and the expression of HGF and the activation of MET were assessed through RT-qPCR or western blot analyses. The mRNA levels of HGF in both U87MG and U87MGR cells were increased upon TMZ treatment (Fig. 2H and I). However, western blot results were inconsistent, possibly due to protein secretion into the extracellular space. The expression levels of MET in U87MG cells were reduced markedly in a dose-dependent manner. However, in U87MGR cells, MET expression was maintained, and its phosphorylation level was elevated with increasing doses of TMZ (Fig. 2J). These results revealed increased HGF expression and secretion in TMZ-resistant GBM cells, accompanied by enhanced MET receptor activation. Even under adverse conditions such as starvation or drug treatment, resistant cell lines maintained elevated HGF and MET expression. This strongly implies an association between HGF/MET pathway activation and TMZ resistance.
Lidocaine enhances TMZ cytotoxicity and reduces cell mobility, potentially by decreasing MET pathway activity in U87MG cells
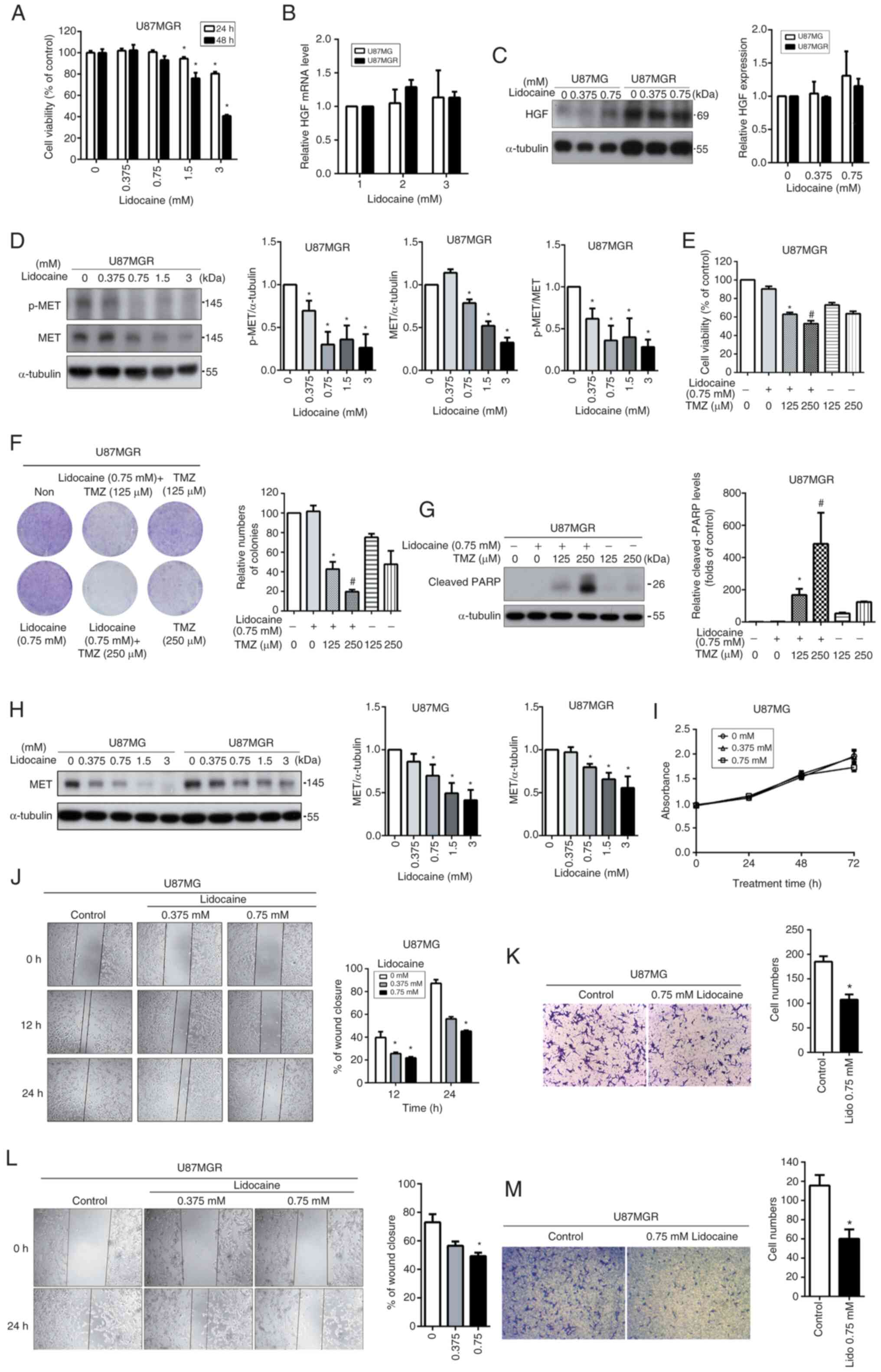
A recent study demonstrated that lidocaine can reduce the mobility of cancer cells and enhance their sensitivity to drug treatment by inhibiting the MET pathway (35). Given the increased activation of the MET pathway observed in TMZ-resistant GBM cells, it was reasoned that if lidocaine can inhibit the MET pathway, it may potentially suppress the malignant behavior of GBM cells and enhance the therapeutic effect of TMZ. To evaluate the cytotoxic effect of lidocaine, U87MGR cells were treated with lidocaine (0–3 mM) for 24 and 48 h, and cell viability was assessed using the MTT assay. Lidocaine significantly reduced cell viability only at concentrations higher than 1.5 mM (Fig. 3A). Subsequently, it was investigated whether lidocaine would impact the expression levels of HGF. Lidocaine did not have a substantial impact on HGF expression in both U87MG and U87MGR cells (Fig. 3B and C). The effect of lidocaine on the MET pathway was further analyzed using western blotting. Lidocaine effectively reduced the expression and activation of MET in a dose-dependent manner, even at a non-toxic concentration of 0.75 mM (Fig. 3D). Next, U87MGR cells were treated with a non-toxic dose of lidocaine, and then stimulated with TMZ for combined treatment to observe whether TMZ sensitivity could be restored in TMZ-resistant cells. Both the MTT assay (Fig. 3E) and colony formation assay (Fig. 3F) indicated that TMZ combined with 0.75 mM lidocaine could effectively enhance TMZ sensitivity in U87MGR cells. The trend observed in the cleaved PARP (Fig. 3G) was consistent with the results from the cell function assays. Based on the aforementioned experimental findings, lidocaine was found to primarily inhibit the expression of MET in U87MGR cells. The effect of lidocaine on the expression of MET was also compared between U87MG and U87MGR cells. U87MGR cells exhibited higher levels of MET expression compared with U87MG cells (Fig. 3H). However, both cell types showed a dose-dependent decrease in MET expression in response to lidocaine treatment. Thus, lidocaine primarily attenuates the activation intensity of the MET pathway by downregulating MET expression levels, rather than impacting the expression of HGF. Taken together, lidocaine cannot directly reverse the high expression of HGF and dephosphorylate MET in U87MGR cells. However, by reducing the expression of MET, it effectively diminishes the overall phosphorylation levels of MET in U87MGR cells. In addition, the effects of lidocaine on cell growth and migration in U87MG cells were further investigated. Lidocaine did not significantly inhibit the growth of U87MG cells, with only a slight inhibition observed after 72 h of treatment (Fig. 3I). To determine whether lidocaine affects cell motility in U87MG and U87MGR cells, the cells were treated with non-toxic concentrations of lidocaine (0.375–0.75 mM) and cell migration was measured through wound healing (Fig. 3J and L) and Transwell assays (Fig. 3K and M). The results revealed that lidocaine significantly inhibited cell migration in both cell types. These results indicated that the effects of lidocaine on TMZ sensitivity and migration are likely due to a decrease in the activity of the MET pathway in the U87MG cell line.
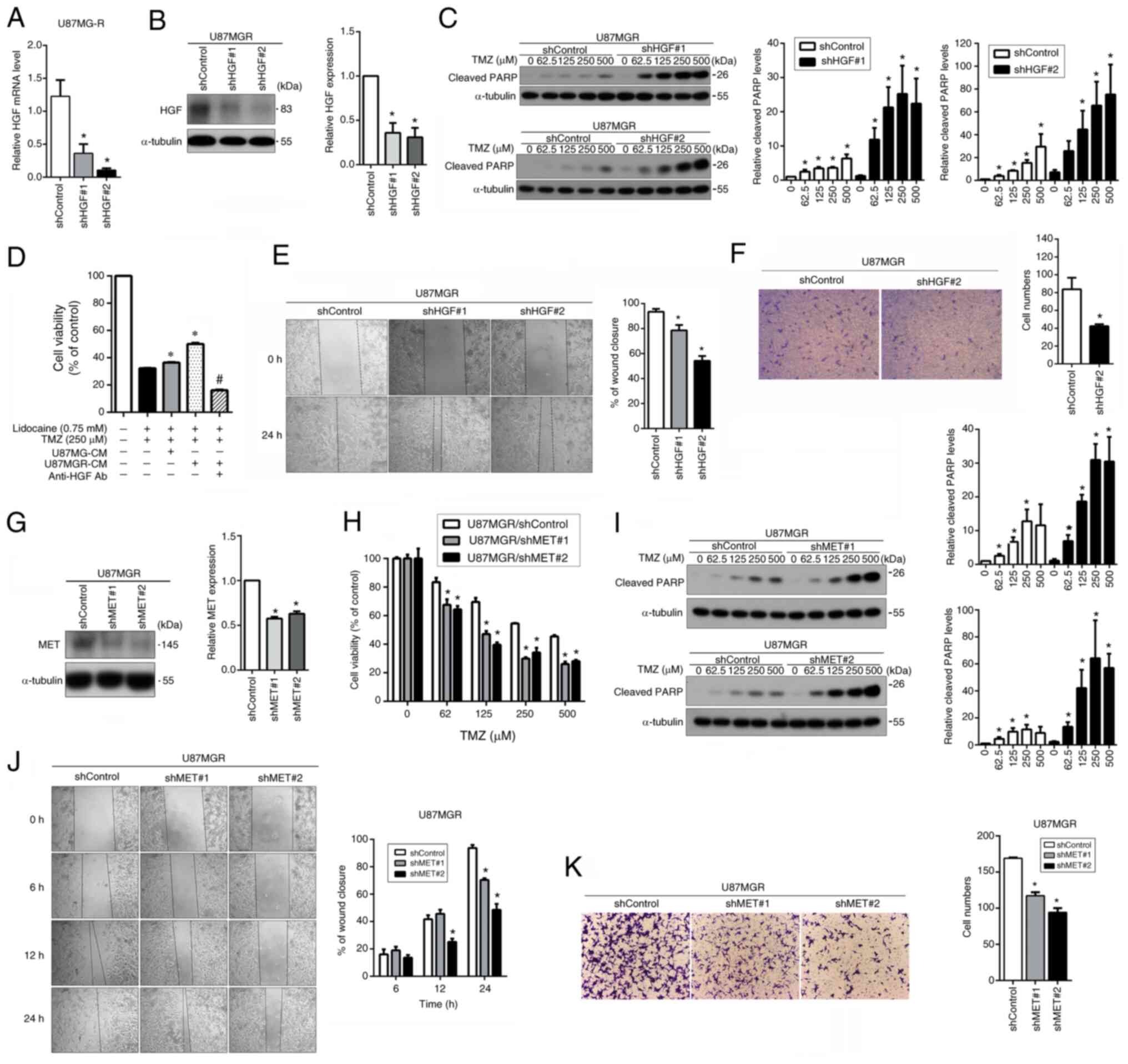
Inhibition of the HGF/MET pathway improves the effectiveness of TMZ and hinders cell migration in U87MGR cells
Since lidocaine can reduce the expression and activation of MET, which affects cellular functions, a gene knockdown experiment was conducted for verification. First, the expression of HGF in U87MGR cells was knocked down using a lentiviral system, and its inhibitory effect was examined through RT-qPCR and western blotting. The expression of HGF in the cells was significantly suppressed (Fig. 4A and B). Western blot analysis of cleaved PARP levels revealed that reducing HGF expression in U87MGR cells substantially elevated TMZ sensitivity (Fig. 4C). Due to the higher expression and secretion of HGF in U87MGR cells compared with U87MG cells, their respective CM were collected. Then, U87MG cells were pre-treated with the two types of concentrated CM and their effect on the combined treatment efficacy of lidocaine and TMZ was analyzed. The CM from U87MGR cells significantly increased the resistance of U87MG cells to TMZ. However, when HGF antibodies were used to react with HGF in the CM from U87MGR cells first, the sensitivity of U87MG cells to TMZ was effectively restored (Fig. 4D). These results indicated that activation of the HGF/MET pathway may induce TMZ resistance, and lidocaine can improve TMZ sensitivity by inhibiting this pathway. Subsequently, it was investigated whether the migration capability of U87MGR cells was affected by the downregulation of HGF gene expression. Suppressed HGF expression significantly inhibited cell migration, as assessed by the wound healing assay (Fig. 4E) and Transwell assay (Fig. 4F). MET expression was also knocked down, which resulted in a strong inhibition of MET expression (Fig. 4G). MTT and western blot analysis demonstrated that downregulation of MET expression significantly enhanced TMZ sensitivity in U87MGR cells (Fig. 4H and I). Next, the impact of MET knockdown on cell mobility was evaluated through wound healing and Transwell assays. Reduced MET expression significantly impaired cell migration (Fig. 4J and K). These results suggested that inhibiting the HGF/MET pathway can effectively enhance TMZ sensitivity and reduce mobility in U87MGR cells.
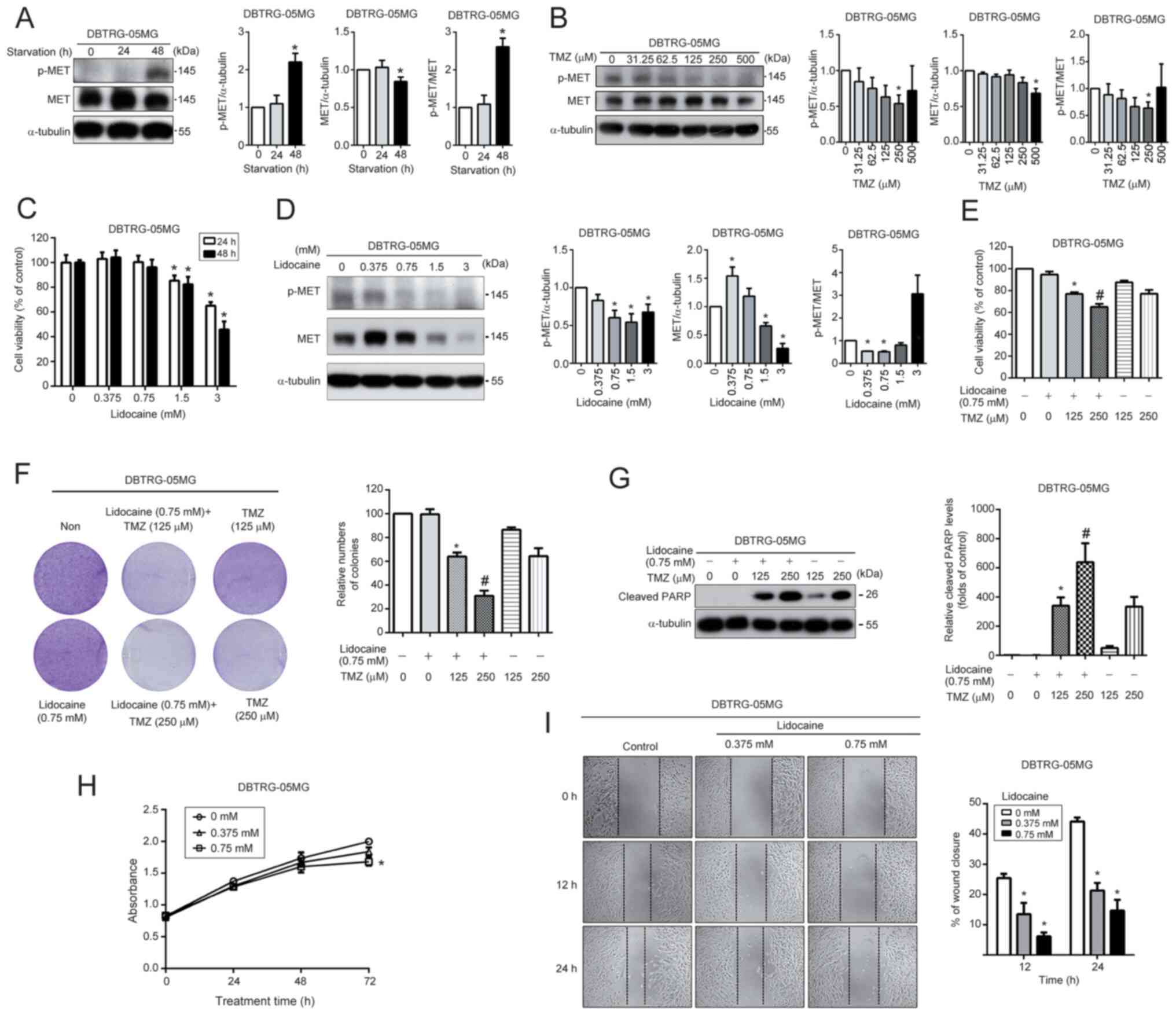
In DBTRG-05MG cells, lidocaine can enhance TMZ sensitivity and reduce cell migration by inhibiting HGF/MET pathway activity
To verify the present findings, another GBM cell line, DBTRG-05MG cells, were selected to investigate the effect of lidocaine on the MET pathway and further analyze its relationship with TMZ sensitivity and cell migration. This choice was based on a previous study by the authors, which revealed that DBTRG-05MG cells exhibit a high level of resistance to TMZ (37). The DBTRG-05MG cells were cultured in a serum-free medium for 24 and 48 h, and the impact of starvation on MET activation was evaluated using western blotting. The MET pathway was significantly activated in cells after 48 h of starvation (Fig. 5A). Furthermore, TMZ was able to significantly inhibit MET activation in a dose-dependent manner (Fig. 5B). The cytotoxicity of lidocaine was evaluated by exposing DBTRG-05MG cells to various concentrations of lidocaine (0–3 mM) for 24 and 48 h, and cell viability was assessed using the MTT assay. Lidocaine significantly decreased cell viability only at concentrations higher than 1.5 mM (Fig. 5C). Furthermore, lidocaine effectively reduced the expression and activation of MET in a dose-dependent manner (Fig. 5D). After co-treatment with 0.75 mM lidocaine and TMZ (125–250 µM) for 48 h, DBTRG-05MG cells exhibited an increase in TMZ sensitivity compared with treatment with TMZ alone, as determined using the MTT assay (Fig. 5E), colony formation (Fig. 5F), and analysis of cleaved PARP levels (Fig. 5G). The effect of lidocaine on the growth and migration of DBTRG-05MG cells was also analyzed. A slight inhibition was observed only after treating cells with lidocaine for 72 h (Fig. 5H). In addition, lidocaine was found to have a significant ability to reduce cell migration (Fig. 5I). These results suggested that lidocaine can enhance TMZ sensitivity and inhibit cell migration by suppressing the activation of the HGF/MET signaling pathway also in DBTRG-05MG cells.
Discussion
In the present study, bioinformatics tools were used to analyze the expression of HGF and MET, as well as the phosphorylation level of MET, in clinical tissue samples to investigate the association between these factors and clinical deterioration and patient prognosis. TMZ-resistant cell lines were also established and it was found that the HGF/MET pathway was highly activated in these cells. However, lidocaine treatment effectively reduced this activation. The present findings indicated that lidocaine not only restores the sensitivity of TMZ-resistant GBM cells to TMZ, but also significantly suppresses cell migration. Furthermore, knockdown of HGF and MET resulted in a significant enhancement of TMZ sensitivity and reduction in cell migration. Overall, these data suggested that lidocaine may enhance TMZ sensitivity and reduce cell migration by inhibiting the activation of the HGF/MET pathway.
In a previous study, lidocaine was revealed to reduce the malignant behavior of gastric cancer cells by inhibiting the c-Met pathway (35). However, the effect of lidocaine on the HGF/MET pathway in GBM cells remains unclear. Through bioinformatics analysis of clinical samples from brain cancer, an association was identified between the activation of the HGF/MET pathway and the malignancy of GBM. Subsequent experiments demonstrated that lidocaine enhances TMZ sensitivity in U87MGR cells by reducing the activity of the HGF/MET pathway, concurrently decreasing cell migration. The current observations indicated that lidocaine primarily acts by decreasing the expression of MET rather than HGF, effectively inhibiting the activation of the HGF/MET pathway. In GBM, the actual activation of MET appears to have a more significant impact on patient prognosis than its expression level. The authors' research has unveiled that the primary effect of lidocaine is to inhibit the expression of MET, contributing to the suppression of aberrant HGF/MET axis activation. The present study provides preliminary insights into the regulatory role of lidocaine on the HGF/MET pathway in GBM, offering potential new directions for further research and clinical treatment. These findings have not been reported previously in the existing literature (35).
Lidocaine, a widely used local anesthetic agent, not only possesses anesthetic properties but also exhibits potential off-target cytotoxic effects that could potentially be utilized as an anticancer treatment. An increasing number of studies suggests that lidocaine can enhance the efficacy of chemotherapy drugs in cancer treatment. Studies have reported that lidocaine has the potential to serve as a valuable adjuvant in cancer treatment. For instance, it has been found to sensitize the cytotoxicity of 5-fluorouracil in melanoma cells (25), enhance the effects of chemotherapeutic drugs against bladder cancer (26), and increase the effectiveness of palbociclib in triple-negative breast cancer (38).
Additionally, several studies have confirmed that lidocaine has the ability to alleviate drug resistance in cancer cells. It has been found to suppress cisplatin resistance in cutaneous squamous cell carcinoma (39), alleviate resistance of gastric cancer cells to cisplatin (29), and reduce resistance of lung cancer cells to cisplatin (28). These findings highlight the potential of lidocaine as a potent agent in reversing drug resistance in various types of cancer. However, as of now, there have been no studies on whether lidocaine enhances the sensitivity of GBM cells to TMZ.
In the past few years, multiple in vivo studies have confirmed the excellent anticancer activity of lidocaine. A study using an in vivo xenograft mouse model has shown that lidocaine not only has the ability to inhibit tumor growth but also significantly enhances the effect of cisplatin chemotherapy (23). Previous literature has also demonstrated that lidocaine effectively reduces tumor growth in a mouse model of melanoma where the tumors were subcutaneously implanted (40). Likewise, numerous studies have reported that exposure to lidocaine reduces tumor growth in mice with subcutaneously implanted human ovarian cancer cells, demonstrating its effectiveness in inhibiting tumor growth (22,41). In recent years, the potential application of lidocaine in clinical cancer treatment has also been discussed, but more clinical data are needed to confirm its efficacy.
The administration of lidocaine may result in adverse effects on the heart, such as arrhythmia, hypotension, or heart failure, particularly in patients with cardiac diseases (42). The recommended dose of lidocaine for managing postoperative pain is 1–2 mg/kg (43). A previous study has found that in colorectal surgery, intravenous administration of lidocaine at a dose of 1.5 mg/kg resulted in an average plasma concentration of 4.0 µg/ml (range, 0.6–12.3 µg/ml, equivalent to 2.56–52.49 µM) (44). At these concentrations, no adverse events or symptoms of local anesthetic toxicity were observed. The lidocaine concentration used in the present study was 0.75 mM, which is higher than the commonly used concentration in clinical practice. Therefore, further preclinical and clinical studies are needed to support its use in cancer treatment. However, by administering through local injection rather than intravenous infusion, it may be possible to increase the local concentration and enhance the potential for combination therapy. On the other hand, lidocaine can cross the blood-brain barrier (45) and reach the brain, making it highly suitable for treating GBM. The current maximum safe dose of lidocaine for in vitro anticancer treatment is 1 mM (46). To enhance its antitumor activity and reduce potential side effects, researchers have explored the use of encapsulation for drug delivery, which could increase the possibility of clinical application (47,48).
Beyond its ability to alleviate surgical pain and eradicate cancer cells, lidocaine also has the potential to inhibit immune-inflammatory responses. Retrospective studies have shown that lidocaine improves survival in patients with pancreatic (49) and ovarian cancer (50). Recent research has revealed that lidocaine can improve the early quality of recovery after brain tumor resection under general anesthesia due to its anti-inflammatory and analgesic properties (51), suggesting that lidocaine may have the potential to enhance the postoperative outcome of GBM surgery.
There is accumulating evidence suggesting that the HGF/MET axis plays a critical role in various aspects of glioblastoma cell behavior, including proliferation (52), growth (53), angiogenesis (54), migration (55), invasion (56), therapeutic resistance (57) and stem cell characteristics (58). The HGF/MET pathway, which plays a critical role in the malignancy of glioblastoma cells, holds great promise as a target for therapeutic intervention. However, there has been a lack of research on the impact of lidocaine on its regulation in GBM cells. The present study, on the other hand, represents a pioneering investigation in this field, as it conclusively demonstrates that lidocaine effectively suppresses the activation of the HGF/MET pathway in GBM cells.
In conclusion, the present study suggests that the HGF/MET pathway plays a critical role in the progression of brain cancer, particularly in GBM. Activation of this pathway in GBM was found to be associated with increased malignancy and poorer patient outcomes. Additionally, it was observed that elevated levels of HGF and activation of its receptor were associated with TMZ resistance. Lidocaine can effectively inhibit the HGF/MET pathway and restore TMZ sensitivity in TMZ-resistant cells. Not only did lidocaine boost the effectiveness of TMZ, but it also demonstrated the capability to impede cell migration. The findings of the present study suggested that the inhibition of the HGF/MET pathway by lidocaine may offer a promising new approach for addressing GBM and enhancing patient outcomes.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Ditmanson Medical Foundation Chia-Yi Christian Hospital (grant no. R110-059).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the OMIX repository (https://ngdc.cncb.ac.cn/omix/release/OMIX005831). The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MSC and JCC conceived and designed the study. ZYC and PHS performed the experiments. CH and HCH were responsible for data analysis and interpretation. MSC and JCC drafted the manuscript. All authors have read and approved the final manuscript. MSC and JCC confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, et al: The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol. 23:1231–1251. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS: CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 15 (Suppl 2):ii1–56. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al: Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Koshy M, Villano JL, Dolecek TA, Howard A, Mahmood U, Chmura SJ, Weichselbaum RR and McCarthy BJ: Improved survival time trends for glioblastoma using the SEER 17 population-based registries. J Neurooncol. 107:207–212. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Fu J, Su X, Li Z, Deng L, Liu X, Feng X and Peng J: HGF/c-MET pathway in cancer: From molecular characterization to clinical evidence. Oncogene. 40:4625–4651. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Trusolino L, Bertotti A and Comoglio PM: MET signalling: Principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 11:834–848. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Mulcahy EQX, Colόn RR and Abounader R: HGF/MET signaling in malignant brain tumors. Int J Mol Sci. 21:75462020. View Article : Google Scholar : PubMed/NCBI | |
|
Arrieta O, Garcia E, Guevara P, Garcia-Navarrete R, Ondarza R, Rembao D and Sotelo J: Hepatocyte growth factor is associated with poor prognosis of malignant gliomas and is a predictor for recurrence of meningioma. Cancer. 94:3210–3218. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Garcia-Navarrete R, Garcia E, Arrieta O and Sotelo J: Hepatocyte growth factor in cerebrospinal fluid is associated with mortality and recurrence of glioblastoma, and could be of prognostic value. J Neurooncol. 97:347–351. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Liu W, Fu Y, Xu S, Ding F, Zhao G, Zhang K, Du C, Pang B and Pang Q: c-Met expression is associated with time to recurrence in patients with glioblastoma multiforme. J Clin Neurosci. 18:119–121. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Olmez OF, Cubukcu E, Evrensel T, Kurt M, Avci N, Tolunay S, Bekar A, Deligonul A, Hartavi M, Alkis N and Manavoglu O: The immunohistochemical expression of c-Met is an independent predictor of survival in patients with glioblastoma multiforme. Clin Transl Oncol. 16:173–177. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Petterson SA, Dahlrot RH, Hermansen SK, KA Munthe S, Gundesen MT, Wohlleben H, Rasmussen T, Beier CP, Hansen S and Kristensen BW: High levels of c-Met is associated with poor prognosis in glioblastoma. J Neurooncol. 122:517–527. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Tabouret E, Denicolai E, Delfino C, Graillon T, Boucard C, Nanni I, Padovani L, Figarella-Branger D and Chinot O: Changes in PlGF and MET-HGF expressions in paired initial and recurrent glioblastoma. J Neurooncol. 130:431–437. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Theofanis T, Chalouhi N, Dalyai R, Starke RM, Jabbour P, Rosenwasser RH and Tjoumakaris S: Microsurgery for cerebral arteriovenous malformations: postoperative outcomes and predictors of complications in 264 cases. Neurosurg Focus. 37:E102014. View Article : Google Scholar : PubMed/NCBI | |
|
Hu H, Mu Q, Bao Z, Chen Y, Liu Y, Chen J, Wang K, Wang Z, Nam Y, Jiang B, et al: Mutational landscape of secondary glioblastoma guides MET-Targeted trial in brain tumor. Cell. 175:1665–1678.e18. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Hermanns H, Hollmann MW, Stevens MF, Lirk P, Brandenburger T, Piegeler T and Werdehausen R: Molecular mechanisms of action of systemic lidocaine in acute and chronic pain: A narrative review. Br J Anaesth. 123:335–349. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang C, Xie C and Lu Y: Local anesthetic lidocaine and cancer: Insight into tumor progression and recurrence. Front Oncol. 11:6697462021. View Article : Google Scholar : PubMed/NCBI | |
|
Sakaguchi M, Kuroda Y and Hirose M: The antiproliferative effect of lidocaine on human tongue cancer cells with inhibition of the activity of epidermal growth factor receptor. Anesth Analg. 102:1103–1107. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Hirata M, Sakaguchi M, Mochida C, Sotozono C, Kageyama K, Kuroda Y and Hirose M: Lidocaine inhibits tyrosine kinase activity of the epidermal growth factor receptor and suppresses proliferation of corneal epithelial cells. Anesthesiology. 100:1206–1210. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Sun H and Sun Y: Lidocaine inhibits proliferation and metastasis of lung cancer cell via regulation of miR-539/EGFR axis. Artif Cells Nanomed Biotechnol. 47:2866–2874. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Sui H, Lou A, Li Z and Yang J: Lidocaine inhibits growth, migration and invasion of gastric carcinoma cells by up-regulation of miR-145. BMC Cancer. 19:2332019. View Article : Google Scholar : PubMed/NCBI | |
|
Sun D, Li YC and Zhang XY: Lidocaine promoted ferroptosis by targeting miR-382-5p/SLC7A11 axis in ovarian and breast cancer. Front Pharmacol. 12:6812232021. View Article : Google Scholar : PubMed/NCBI | |
|
Xing W, Chen DT, Pan JH, Chen YH, Yan Y, Li Q, Xue RF, Yuan YF and Zeng WA: Lidocaine Induces apoptosis and suppresses tumor growth in human hepatocellular carcinoma cells in vitro and in a xenograft model in vivo. Anesthesiology. 126:868–881. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Mirshahidi S, Shields TG, de Necochea-Campion R, Yuan X, Janjua A, Williams NL, Mirshahidi HR, Reeves ME, Duerksen-Hughes P and Zuckerman LM: Bupivacaine and lidocaine induce apoptosis in osteosarcoma tumor cells. Clin Orthop Relat Res. 479:180–194. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Y, Xie J, Liu W, Zhang R, Huang S and Xing Y: Lidocaine sensitizes the cytotoxicity of 5-fluorouacil in melanoma cells via upregulation of microRNA-493. Pharmazie. 72:663–669. 2017.PubMed/NCBI | |
|
Yang X, Zhao L, Li M, Yan L, Zhang S, Mi Z, Ren L and Xu J: Lidocaine enhances the effects of chemotherapeutic drugs against bladder cancer. Sci Rep. 8:5982018. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang X, Pang W, Liu H and Wang J: Lidocine potentiates the cytotoxicity of 5-fluorouracil to choriocarcinoma cells by downregulating ABC transport proteins expression. J Cell Biochem. 120:16533–16542. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Yang Q, Zhang Z, Xu H and Ma C: Lidocaine alleviates cytotoxicity-resistance in lung cancer A549/DDP cells via down-regulation of miR-21. Mol Cell Biochem. 456:63–72. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang X, Gu G, Li X and Zhang C: Lidocaine alleviates cisplatin resistance and inhibits migration of MGC-803/DDP cells through decreasing miR-10b. Cell Cycle. 19:2530–2537. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Wen J, Li X, Ding Y, Zheng S and Xiao Y: Lidocaine inhibits glioma cell proliferation, migration and invasion by modulating the circEZH2/miR-181b-5p pathway. Neuroreport. 32:52–60. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Li D, Yang X, Li B, Yang C, Sun J, Yu M, Wang H and Lu Y: Lidocaine liposome modified with folic acid suppresses the proliferation and motility of glioma cells via targeting the PI3K/AKT pathway. Exp Ther Med. 22:10252021. View Article : Google Scholar : PubMed/NCBI | |
|
Leng T, Lin S, Xiong Z and Lin J: Lidocaine suppresses glioma cell proliferation by inhibiting TRPM7 channels. Int J Physiol Pathophysiol Pharmacol. 9:8–15. 2017.PubMed/NCBI | |
|
Lu J, Ju YT, Li C, Hua FZ, Xu GH and Hu YH: Effect of TRPV1 combined with lidocaine on cell state and apoptosis of U87-MG glioma cell lines. Asian Pac J Trop Med. 9:288–292. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Fan X, Yang H, Zhao C, Hu L, Wang D, Wang R, Fang Z and Chen X: Local anesthetics impair the growth and self-renewal of glioblastoma stem cells by inhibiting ZDHHC15-mediated GP130 palmitoylation. Stem Cell Res Ther. 12:1072021. View Article : Google Scholar : PubMed/NCBI | |
|
Zeng W, Xing ZT, Tan MY, Wu YW and Zhang CY: Lidocaine suppresses the malignant behavior of gastric cancer cells via the c-Met/c-Src pathway. Exp Ther Med. 21:4242021. View Article : Google Scholar : PubMed/NCBI | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Yang JT, Lee IN, Huang C, Huang HC, Wu YP, Chong ZY and Chen JC: ADAM17 confers temozolomide resistance in human glioblastoma cells and miR-145 regulates its expression. Int J Mol Sci. 24:77032023. View Article : Google Scholar : PubMed/NCBI | |
|
Han BS, Jung KH, Lee JE, Yoon YC, Ko S, Park MS, Lee YJ, Kim SE, Cho YJ, Lee P, et al: Lidocaine enhances the efficacy of palbociclib in triple-negative breast cancer. Am J Cancer Res. 12:3083–3098. 2022.PubMed/NCBI | |
|
Liu T, Jiang F, Yu LY and Wu YY: Lidocaine represses proliferation and cisplatin resistance in cutaneous squamous cell carcinoma via miR-30c/SIRT1 regulation. Bioengineered. 13:6359–6370. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Chen J, Jiao Z, Wang A and Zhong W: Lidocaine inhibits melanoma cell proliferation by regulating ERK phosphorylation. J Cell Biochem. 120:6402–6408. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Gao R, Cao T, Chen H, Cai J, Lei M and Wang Z: Nav1.5-E3 antibody inhibits cancer progression. Transl Cancer Res. 8:44–50. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Tierney KJ, Murano T and Natal B: Lidocaine-Induced cardiac arrest in the emergency department: Effectiveness of lipid therapy. J Emerg Med. 50:47–50. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Chu R, Umukoro N, Greer T, Roberts J, Adekoya P, Odonkor CA, Hagedorn JM, Olatoye D, Urits I, Orhurhu MS, et al: Intravenous lidocaine infusion for the management of early postoperative pain: A comprehensive review of controlled trials. Psychopharmacol Bull. 50 (4 Suppl 1):S216–S259. 2020. | |
|
Greenwood E, Nimmo S, Paterson H, Homer N and Foo I: Intravenous lidocaine infusion as a component of multimodal analgesia for colorectal surgery-measurement of plasma levels. Perioper Med (Lond). 8:12019. View Article : Google Scholar : PubMed/NCBI | |
|
Beaussier M, Delbos A, Maurice-Szamburski A, Ecoffey C and Mercadal L: Perioperative use of intravenous lidocaine. Drugs. 78:1229–1246. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Li K, Yang J and Han X: Lidocaine sensitizes the cytotoxicity of cisplatin in breast cancer cells via up-regulation of RARβ2 and RASSF1A demethylation. Int J Mol Sci. 15:23519–23536. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Gao X, Yang H, Wu M, Shi K, Zhou C, Peng J and Yang Q: Targeting delivery of lidocaine and cisplatin by nanogel enhances chemotherapy and alleviates metastasis. ACS Appl Mater Interfaces. 10:25228–25240. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Yang Y, Sun J, Peng F, Liu H, Zhao G, Chen J, Zhang W and Qiu F: Enhanced antitumor activity of lidocaine nanoparticles encapsulated by a self-assembling peptide. Front Pharmacol. 13:7708922022. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang H, Yang L, Zhu X, Zhu M, Sun Z, Cata JP, Chen W and Miao C: Association between intraoperative intravenous lidocaine infusion and survival in patients undergoing pancreatectomy for pancreatic cancer: A retrospective study. Br J Anaesth. 125:141–148. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang H, Gu J, Qu M, Sun Z, Huang Q, Cata JP, Chen W and Miao C: Effects of intravenous infusion of lidocaine on short-term outcomes and survival in patients undergoing surgery for ovarian cancer: A retrospective propensity score matching study. Front Oncol. 11:6898322022. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao K, Dong Y, Su G, Wang Y, Ji T, Wu N, Cui X, Li W, Yang Y and Chen X: Effect of systemic lidocaine on postoperative early recovery quality in patients undergoing supratentorial tumor resection. Drug Des Devel Ther. 16:1171–1181. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Yamamoto S, Wakimoto H, Aoyagi M, Hirakawa K and Hamada H: Modulation of motility and proliferation of glioma cells by hepatocyte growth factor. Jpn J Cancer Res. 88:564–577. 1997. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao Y, Sun Y, Zhang H, Liu X, Du W, Li Y, Zhang J, Chen L and Jiang C: HGF/MET signaling promotes glioma growth via up-regulation of Cox-2 expression and PGE2 production. Int J Clin Exp Pathol. 8:3719–3726. 2015.PubMed/NCBI | |
|
Laterra J, Nam M, Rosen E, Rao JS, Lamszus K, Goldberg ID and Johnston P: Scatter factor/hepatocyte growth factor gene transfer enhances glioma growth and angiogenesis in vivo. Lab Invest. 76:565–577. 1997.PubMed/NCBI | |
|
Eckerich C, Zapf S, Fillbrandt R, Loges S, Westphal M and Lamszus K: Hypoxia can induce c-Met expression in glioma cells and enhance SF/HGF-induced cell migration. Int J Cancer. 121:276–283. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Li W, Xiong X, Abdalla A, Alejo S, Zhu L, Lu F and Sun H: HGF-induced formation of the MET-AXL-ELMO2-DOCK180 complex promotes RAC1 activation, receptor clustering, and cancer cell migration and invasion. J Biol Chem. 293:15397–15418. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Jun HJ, Acquaviva J, Chi D, Lessard J, Zhu H, Woolfenden S, Bronson RT, Pfannl R, White F, Housman DE, et al: Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene. 31:3039–3050. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Casey BJ, Somerville LH, Gotlib IH, Ayduk O, Franklin NT, Askren MK, Jonides J, Berman MG, Wilson NL, Teslovich T, et al: Behavioral and neural correlates of delay of gratification 40 years later. Proc Natl Acad Sci USA. 108:14998–15003. 2011. View Article : Google Scholar : PubMed/NCBI |